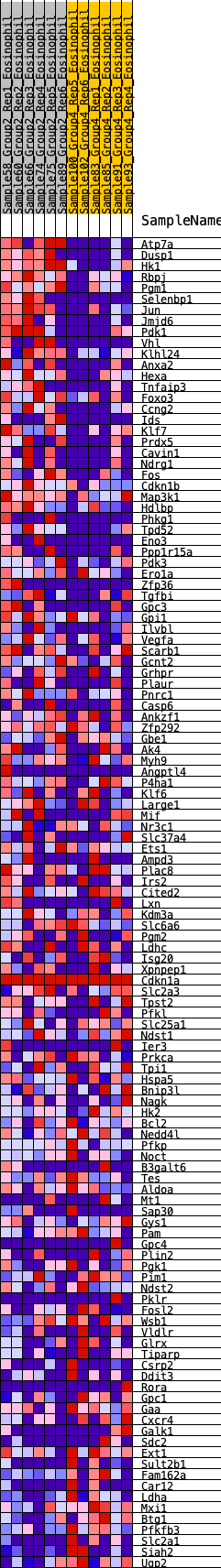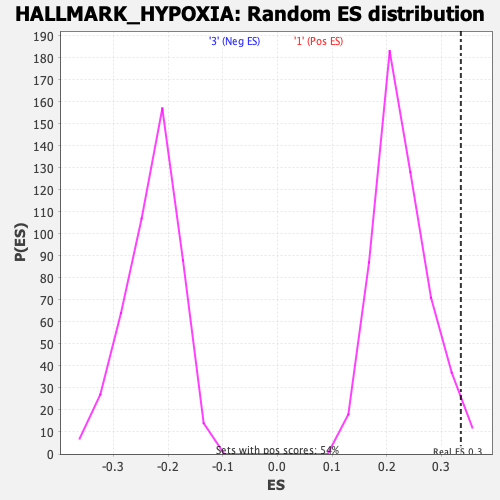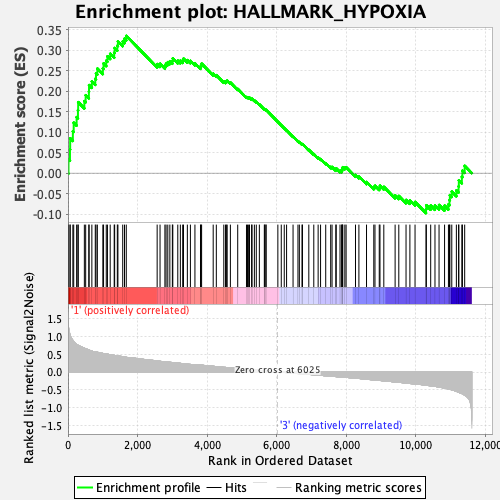
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.33542314 |
| Normalized Enrichment Score (NES) | 1.4775391 |
| Nominal p-value | 0.029850746 |
| FDR q-value | 0.116116956 |
| FWER p-Value | 0.354 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp7a | 15 | 1.298 | 0.0336 | Yes |
| 2 | Dusp1 | 57 | 1.045 | 0.0582 | Yes |
| 3 | Hk1 | 63 | 1.019 | 0.0851 | Yes |
| 4 | Rbpj | 140 | 0.885 | 0.1023 | Yes |
| 5 | Pgm1 | 164 | 0.854 | 0.1233 | Yes |
| 6 | Selenbp1 | 248 | 0.774 | 0.1368 | Yes |
| 7 | Jun | 286 | 0.753 | 0.1539 | Yes |
| 8 | Jmjd6 | 293 | 0.747 | 0.1734 | Yes |
| 9 | Pdk1 | 470 | 0.667 | 0.1760 | Yes |
| 10 | Vhl | 508 | 0.653 | 0.1904 | Yes |
| 11 | Klhl24 | 600 | 0.616 | 0.1990 | Yes |
| 12 | Anxa2 | 605 | 0.614 | 0.2152 | Yes |
| 13 | Hexa | 686 | 0.584 | 0.2240 | Yes |
| 14 | Tnfaip3 | 784 | 0.563 | 0.2307 | Yes |
| 15 | Foxo3 | 805 | 0.560 | 0.2440 | Yes |
| 16 | Ccng2 | 844 | 0.553 | 0.2555 | Yes |
| 17 | Ids | 1002 | 0.521 | 0.2559 | Yes |
| 18 | Klf7 | 1022 | 0.517 | 0.2681 | Yes |
| 19 | Prdx5 | 1100 | 0.504 | 0.2750 | Yes |
| 20 | Cavin1 | 1134 | 0.498 | 0.2855 | Yes |
| 21 | Ndrg1 | 1213 | 0.486 | 0.2918 | Yes |
| 22 | Fos | 1328 | 0.467 | 0.2944 | Yes |
| 23 | Cdkn1b | 1341 | 0.465 | 0.3058 | Yes |
| 24 | Map3k1 | 1418 | 0.452 | 0.3114 | Yes |
| 25 | Hdlbp | 1435 | 0.449 | 0.3221 | Yes |
| 26 | Phkg1 | 1573 | 0.430 | 0.3217 | Yes |
| 27 | Tpd52 | 1624 | 0.422 | 0.3287 | Yes |
| 28 | Eno3 | 1675 | 0.411 | 0.3354 | Yes |
| 29 | Ppp1r15a | 2563 | 0.310 | 0.2665 | No |
| 30 | Pdk3 | 2646 | 0.299 | 0.2675 | No |
| 31 | Ero1a | 2786 | 0.285 | 0.2630 | No |
| 32 | Zfp36 | 2819 | 0.283 | 0.2678 | No |
| 33 | Tgfbi | 2869 | 0.280 | 0.2711 | No |
| 34 | Gpc3 | 2928 | 0.272 | 0.2734 | No |
| 35 | Gpi1 | 3000 | 0.265 | 0.2743 | No |
| 36 | Ilvbl | 3013 | 0.265 | 0.2804 | No |
| 37 | Vegfa | 3156 | 0.251 | 0.2748 | No |
| 38 | Scarb1 | 3230 | 0.243 | 0.2750 | No |
| 39 | Gcnt2 | 3298 | 0.235 | 0.2755 | No |
| 40 | Grhpr | 3320 | 0.232 | 0.2799 | No |
| 41 | Plaur | 3431 | 0.220 | 0.2762 | No |
| 42 | Pnrc1 | 3519 | 0.213 | 0.2744 | No |
| 43 | Casp6 | 3649 | 0.201 | 0.2686 | No |
| 44 | Ankzf1 | 3809 | 0.193 | 0.2599 | No |
| 45 | Zfp292 | 3823 | 0.192 | 0.2640 | No |
| 46 | Gbe1 | 3841 | 0.191 | 0.2676 | No |
| 47 | Ak4 | 4174 | 0.161 | 0.2430 | No |
| 48 | Myh9 | 4264 | 0.152 | 0.2394 | No |
| 49 | Angptl4 | 4479 | 0.130 | 0.2243 | No |
| 50 | P4ha1 | 4529 | 0.130 | 0.2235 | No |
| 51 | Klf6 | 4553 | 0.127 | 0.2249 | No |
| 52 | Large1 | 4576 | 0.125 | 0.2263 | No |
| 53 | Mif | 4668 | 0.116 | 0.2215 | No |
| 54 | Nr3c1 | 4879 | 0.096 | 0.2058 | No |
| 55 | Slc37a4 | 5131 | 0.076 | 0.1860 | No |
| 56 | Ets1 | 5161 | 0.073 | 0.1855 | No |
| 57 | Ampd3 | 5193 | 0.070 | 0.1847 | No |
| 58 | Plac8 | 5218 | 0.069 | 0.1844 | No |
| 59 | Irs2 | 5278 | 0.066 | 0.1811 | No |
| 60 | Cited2 | 5291 | 0.065 | 0.1818 | No |
| 61 | Lxn | 5358 | 0.059 | 0.1776 | No |
| 62 | Kdm3a | 5413 | 0.054 | 0.1743 | No |
| 63 | Slc6a6 | 5501 | 0.045 | 0.1680 | No |
| 64 | Pgm2 | 5644 | 0.032 | 0.1565 | No |
| 65 | Ldhc | 5656 | 0.031 | 0.1564 | No |
| 66 | Isg20 | 5691 | 0.028 | 0.1542 | No |
| 67 | Xpnpep1 | 5693 | 0.027 | 0.1548 | No |
| 68 | Cdkn1a | 6033 | 0.000 | 0.1253 | No |
| 69 | Slc2a3 | 6131 | -0.003 | 0.1169 | No |
| 70 | Tpst2 | 6217 | -0.010 | 0.1098 | No |
| 71 | Pfkl | 6284 | -0.015 | 0.1045 | No |
| 72 | Slc25a1 | 6470 | -0.031 | 0.0892 | No |
| 73 | Ndst1 | 6611 | -0.042 | 0.0782 | No |
| 74 | Ier3 | 6651 | -0.046 | 0.0760 | No |
| 75 | Prkca | 6727 | -0.049 | 0.0708 | No |
| 76 | Tpi1 | 6741 | -0.050 | 0.0710 | No |
| 77 | Hspa5 | 6923 | -0.064 | 0.0570 | No |
| 78 | Bnip3l | 7066 | -0.074 | 0.0466 | No |
| 79 | Nagk | 7191 | -0.086 | 0.0381 | No |
| 80 | Hk2 | 7261 | -0.091 | 0.0346 | No |
| 81 | Bcl2 | 7409 | -0.102 | 0.0246 | No |
| 82 | Nedd4l | 7552 | -0.115 | 0.0153 | No |
| 83 | Pfkp | 7591 | -0.117 | 0.0151 | No |
| 84 | Noct | 7700 | -0.126 | 0.0091 | No |
| 85 | B3galt6 | 7710 | -0.127 | 0.0117 | No |
| 86 | Tes | 7820 | -0.136 | 0.0059 | No |
| 87 | Aldoa | 7867 | -0.140 | 0.0057 | No |
| 88 | Mt1 | 7878 | -0.141 | 0.0086 | No |
| 89 | Sap30 | 7889 | -0.142 | 0.0115 | No |
| 90 | Gys1 | 7900 | -0.142 | 0.0145 | No |
| 91 | Pam | 7959 | -0.149 | 0.0134 | No |
| 92 | Gpc4 | 7995 | -0.151 | 0.0145 | No |
| 93 | Plin2 | 8265 | -0.172 | -0.0043 | No |
| 94 | Pgk1 | 8363 | -0.179 | -0.0080 | No |
| 95 | Pim1 | 8582 | -0.200 | -0.0216 | No |
| 96 | Ndst2 | 8791 | -0.219 | -0.0338 | No |
| 97 | Pklr | 8825 | -0.220 | -0.0307 | No |
| 98 | Fosl2 | 8948 | -0.232 | -0.0351 | No |
| 99 | Wsb1 | 8969 | -0.234 | -0.0305 | No |
| 100 | Vldlr | 9079 | -0.245 | -0.0334 | No |
| 101 | Glrx | 9404 | -0.276 | -0.0542 | No |
| 102 | Tiparp | 9510 | -0.286 | -0.0557 | No |
| 103 | Csrp2 | 9718 | -0.307 | -0.0655 | No |
| 104 | Ddit3 | 9829 | -0.319 | -0.0665 | No |
| 105 | Rora | 9977 | -0.332 | -0.0703 | No |
| 106 | Gpc1 | 10293 | -0.368 | -0.0878 | No |
| 107 | Gaa | 10301 | -0.369 | -0.0785 | No |
| 108 | Cxcr4 | 10427 | -0.383 | -0.0791 | No |
| 109 | Galk1 | 10550 | -0.400 | -0.0790 | No |
| 110 | Sdc2 | 10667 | -0.421 | -0.0777 | No |
| 111 | Ext1 | 10826 | -0.450 | -0.0794 | No |
| 112 | Sult2b1 | 10937 | -0.473 | -0.0762 | No |
| 113 | Fam162a | 10967 | -0.479 | -0.0659 | No |
| 114 | Car12 | 10978 | -0.482 | -0.0538 | No |
| 115 | Ldha | 11033 | -0.497 | -0.0451 | No |
| 116 | Mxi1 | 11165 | -0.537 | -0.0421 | No |
| 117 | Btg1 | 11229 | -0.565 | -0.0324 | No |
| 118 | Pfkfb3 | 11236 | -0.568 | -0.0176 | No |
| 119 | Slc2a1 | 11323 | -0.602 | -0.0089 | No |
| 120 | Siah2 | 11339 | -0.607 | 0.0061 | No |
| 121 | Ugp2 | 11404 | -0.644 | 0.0178 | No |