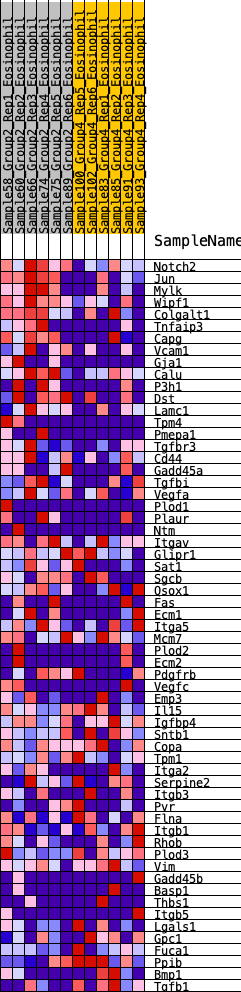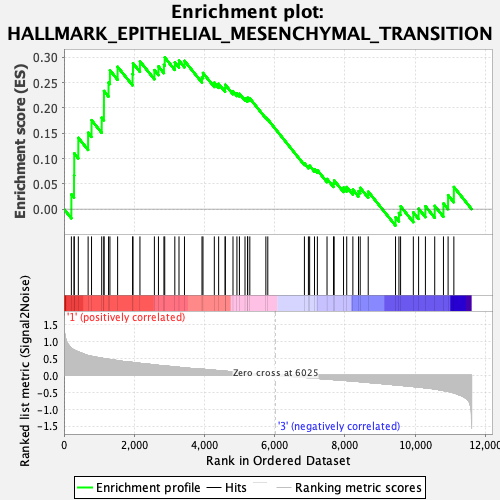
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.30004504 |
| Normalized Enrichment Score (NES) | 1.2327952 |
| Nominal p-value | 0.1378026 |
| FDR q-value | 0.32492745 |
| FWER p-Value | 0.836 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Notch2 | 210 | 0.805 | 0.0291 | Yes |
| 2 | Jun | 286 | 0.753 | 0.0668 | Yes |
| 3 | Mylk | 292 | 0.748 | 0.1103 | Yes |
| 4 | Wipf1 | 408 | 0.693 | 0.1410 | Yes |
| 5 | Colgalt1 | 687 | 0.583 | 0.1512 | Yes |
| 6 | Tnfaip3 | 784 | 0.563 | 0.1759 | Yes |
| 7 | Capg | 1074 | 0.508 | 0.1807 | Yes |
| 8 | Vcam1 | 1137 | 0.497 | 0.2045 | Yes |
| 9 | Gja1 | 1138 | 0.497 | 0.2337 | Yes |
| 10 | Calu | 1269 | 0.475 | 0.2504 | Yes |
| 11 | P3h1 | 1308 | 0.470 | 0.2747 | Yes |
| 12 | Dst | 1527 | 0.437 | 0.2815 | Yes |
| 13 | Lamc1 | 1953 | 0.382 | 0.2671 | Yes |
| 14 | Tpm4 | 1965 | 0.381 | 0.2885 | Yes |
| 15 | Pmepa1 | 2163 | 0.355 | 0.2923 | Yes |
| 16 | Tgfbr3 | 2574 | 0.308 | 0.2749 | Yes |
| 17 | Cd44 | 2690 | 0.295 | 0.2823 | Yes |
| 18 | Gadd45a | 2846 | 0.282 | 0.2855 | Yes |
| 19 | Tgfbi | 2869 | 0.280 | 0.3000 | Yes |
| 20 | Vegfa | 3156 | 0.251 | 0.2900 | No |
| 21 | Plod1 | 3275 | 0.238 | 0.2938 | No |
| 22 | Plaur | 3431 | 0.220 | 0.2933 | No |
| 23 | Ntm | 3930 | 0.184 | 0.2610 | No |
| 24 | Itgav | 3958 | 0.182 | 0.2693 | No |
| 25 | Glipr1 | 4281 | 0.151 | 0.2503 | No |
| 26 | Sat1 | 4404 | 0.138 | 0.2478 | No |
| 27 | Sgcb | 4588 | 0.124 | 0.2393 | No |
| 28 | Qsox1 | 4594 | 0.123 | 0.2461 | No |
| 29 | Fas | 4813 | 0.102 | 0.2332 | No |
| 30 | Ecm1 | 4925 | 0.093 | 0.2291 | No |
| 31 | Itga5 | 4996 | 0.088 | 0.2282 | No |
| 32 | Mcm7 | 5155 | 0.074 | 0.2188 | No |
| 33 | Plod2 | 5228 | 0.068 | 0.2166 | No |
| 34 | Ecm2 | 5231 | 0.068 | 0.2204 | No |
| 35 | Pdgfrb | 5292 | 0.065 | 0.2190 | No |
| 36 | Vegfc | 5749 | 0.023 | 0.1809 | No |
| 37 | Emp3 | 5804 | 0.019 | 0.1773 | No |
| 38 | Il15 | 6847 | -0.059 | 0.0906 | No |
| 39 | Igfbp4 | 6961 | -0.067 | 0.0848 | No |
| 40 | Sntb1 | 6996 | -0.070 | 0.0859 | No |
| 41 | Copa | 7134 | -0.081 | 0.0788 | No |
| 42 | Tpm1 | 7216 | -0.088 | 0.0770 | No |
| 43 | Itga2 | 7491 | -0.109 | 0.0596 | No |
| 44 | Serpine2 | 7675 | -0.124 | 0.0510 | No |
| 45 | Itgb3 | 7694 | -0.125 | 0.0568 | No |
| 46 | Pvr | 7961 | -0.149 | 0.0426 | No |
| 47 | Flna | 8052 | -0.152 | 0.0437 | No |
| 48 | Itgb1 | 8227 | -0.168 | 0.0385 | No |
| 49 | Rhob | 8387 | -0.181 | 0.0354 | No |
| 50 | Plod3 | 8436 | -0.185 | 0.0421 | No |
| 51 | Vim | 8664 | -0.208 | 0.0347 | No |
| 52 | Gadd45b | 9442 | -0.279 | -0.0162 | No |
| 53 | Basp1 | 9539 | -0.288 | -0.0076 | No |
| 54 | Thbs1 | 9586 | -0.292 | 0.0055 | No |
| 55 | Itgb5 | 9949 | -0.329 | -0.0065 | No |
| 56 | Lgals1 | 10099 | -0.344 | 0.0008 | No |
| 57 | Gpc1 | 10293 | -0.368 | 0.0057 | No |
| 58 | Fuca1 | 10558 | -0.401 | 0.0064 | No |
| 59 | Ppib | 10807 | -0.447 | 0.0112 | No |
| 60 | Bmp1 | 10939 | -0.473 | 0.0276 | No |
| 61 | Tgfb1 | 11103 | -0.516 | 0.0438 | No |