Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
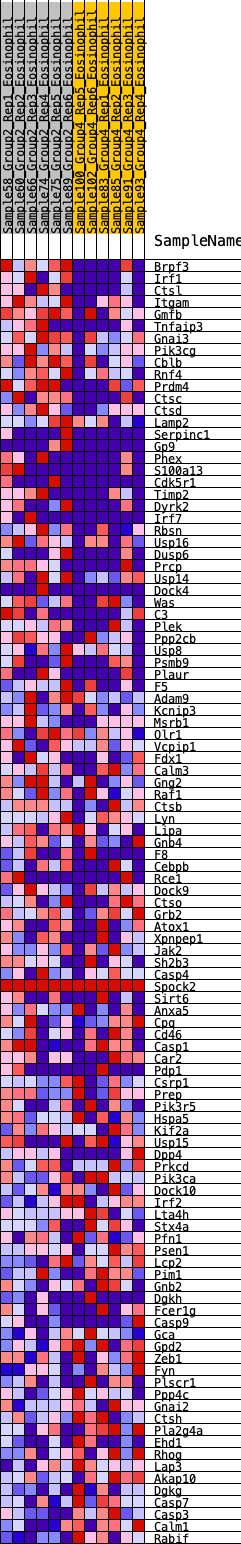
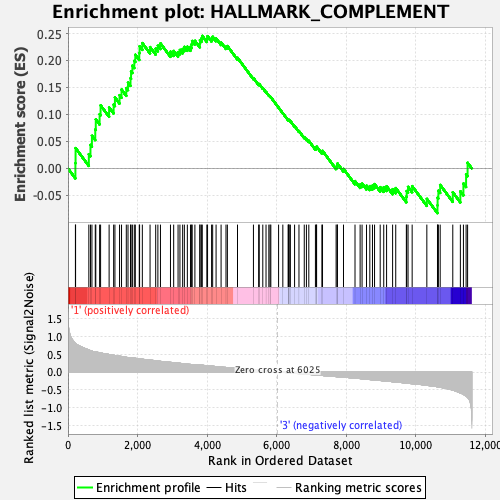
| GeneSet | HALLMARK_COMPLEMENT |
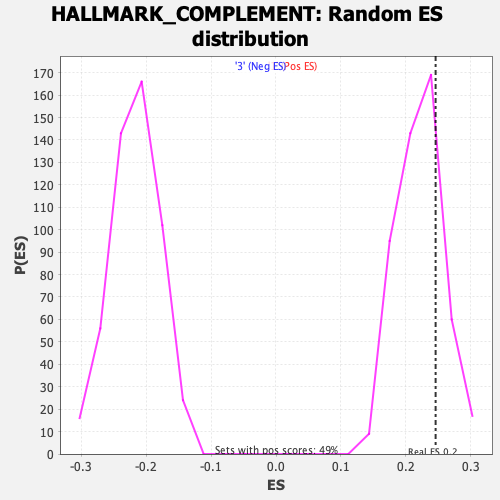
| Enrichment Score (ES) | 0.24559665 |
| Normalized Enrichment Score (NES) | 1.1091241 |
| Nominal p-value | 0.23326571 |
| FDR q-value | 0.5058374 |
| FWER p-Value | 0.971 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Brpf3 | 213 | 0.804 | 0.0099 | Yes |
| 2 | Irf1 | 218 | 0.798 | 0.0377 | Yes |
| 3 | Ctsl | 599 | 0.617 | 0.0264 | Yes |
| 4 | Itgam | 646 | 0.600 | 0.0436 | Yes |
| 5 | Gmfb | 688 | 0.583 | 0.0606 | Yes |
| 6 | Tnfaip3 | 784 | 0.563 | 0.0723 | Yes |
| 7 | Gnai3 | 799 | 0.561 | 0.0909 | Yes |
| 8 | Pik3cg | 909 | 0.539 | 0.1004 | Yes |
| 9 | Cblb | 937 | 0.533 | 0.1169 | Yes |
| 10 | Rnf4 | 1182 | 0.490 | 0.1130 | Yes |
| 11 | Prdm4 | 1312 | 0.469 | 0.1183 | Yes |
| 12 | Ctsc | 1349 | 0.463 | 0.1316 | Yes |
| 13 | Ctsd | 1482 | 0.443 | 0.1357 | Yes |
| 14 | Lamp2 | 1539 | 0.436 | 0.1463 | Yes |
| 15 | Serpinc1 | 1676 | 0.411 | 0.1490 | Yes |
| 16 | Gp9 | 1725 | 0.407 | 0.1592 | Yes |
| 17 | Phex | 1797 | 0.400 | 0.1671 | Yes |
| 18 | S100a13 | 1813 | 0.398 | 0.1799 | Yes |
| 19 | Cdk5r1 | 1851 | 0.392 | 0.1905 | Yes |
| 20 | Timp2 | 1906 | 0.388 | 0.1995 | Yes |
| 21 | Dyrk2 | 1935 | 0.385 | 0.2107 | Yes |
| 22 | Irf7 | 2049 | 0.371 | 0.2140 | Yes |
| 23 | Rbsn | 2058 | 0.370 | 0.2263 | Yes |
| 24 | Usp16 | 2137 | 0.359 | 0.2322 | Yes |
| 25 | Dusp6 | 2359 | 0.334 | 0.2248 | Yes |
| 26 | Prcp | 2521 | 0.316 | 0.2220 | Yes |
| 27 | Usp14 | 2584 | 0.308 | 0.2275 | Yes |
| 28 | Dock4 | 2658 | 0.299 | 0.2317 | Yes |
| 29 | Was | 2946 | 0.271 | 0.2163 | Yes |
| 30 | C3 | 3038 | 0.263 | 0.2177 | Yes |
| 31 | Plek | 3164 | 0.251 | 0.2157 | Yes |
| 32 | Ppp2cb | 3218 | 0.244 | 0.2197 | Yes |
| 33 | Usp8 | 3296 | 0.236 | 0.2213 | Yes |
| 34 | Psmb9 | 3345 | 0.229 | 0.2252 | Yes |
| 35 | Plaur | 3431 | 0.220 | 0.2256 | Yes |
| 36 | F5 | 3521 | 0.213 | 0.2254 | Yes |
| 37 | Adam9 | 3554 | 0.210 | 0.2300 | Yes |
| 38 | Kcnip3 | 3569 | 0.209 | 0.2362 | Yes |
| 39 | Msrb1 | 3648 | 0.201 | 0.2365 | Yes |
| 40 | Olr1 | 3791 | 0.194 | 0.2310 | Yes |
| 41 | Vcpip1 | 3794 | 0.194 | 0.2377 | Yes |
| 42 | Fdx1 | 3835 | 0.191 | 0.2410 | Yes |
| 43 | Calm3 | 3860 | 0.189 | 0.2456 | Yes |
| 44 | Gng2 | 3992 | 0.178 | 0.2405 | No |
| 45 | Raf1 | 4012 | 0.176 | 0.2451 | No |
| 46 | Ctsb | 4128 | 0.165 | 0.2409 | No |
| 47 | Lyn | 4159 | 0.162 | 0.2440 | No |
| 48 | Lipa | 4257 | 0.153 | 0.2410 | No |
| 49 | Gnb4 | 4401 | 0.138 | 0.2334 | No |
| 50 | F8 | 4542 | 0.128 | 0.2258 | No |
| 51 | Cebpb | 4582 | 0.124 | 0.2268 | No |
| 52 | Rce1 | 4873 | 0.096 | 0.2050 | No |
| 53 | Dock9 | 5331 | 0.061 | 0.1674 | No |
| 54 | Ctso | 5482 | 0.047 | 0.1560 | No |
| 55 | Grb2 | 5497 | 0.046 | 0.1564 | No |
| 56 | Atox1 | 5600 | 0.035 | 0.1488 | No |
| 57 | Xpnpep1 | 5693 | 0.027 | 0.1417 | No |
| 58 | Jak2 | 5774 | 0.021 | 0.1355 | No |
| 59 | Sh2b3 | 5808 | 0.018 | 0.1333 | No |
| 60 | Casp4 | 5831 | 0.016 | 0.1320 | No |
| 61 | Spock2 | 6054 | 0.000 | 0.1127 | No |
| 62 | Sirt6 | 6176 | -0.007 | 0.1024 | No |
| 63 | Anxa5 | 6328 | -0.019 | 0.0899 | No |
| 64 | Cpq | 6339 | -0.020 | 0.0898 | No |
| 65 | Cd46 | 6351 | -0.021 | 0.0896 | No |
| 66 | Casp1 | 6356 | -0.021 | 0.0899 | No |
| 67 | Car2 | 6393 | -0.025 | 0.0877 | No |
| 68 | Pdp1 | 6514 | -0.035 | 0.0785 | No |
| 69 | Csrp1 | 6640 | -0.046 | 0.0692 | No |
| 70 | Prep | 6792 | -0.054 | 0.0580 | No |
| 71 | Pik3r5 | 6854 | -0.060 | 0.0548 | No |
| 72 | Hspa5 | 6923 | -0.064 | 0.0512 | No |
| 73 | Kif2a | 7113 | -0.079 | 0.0375 | No |
| 74 | Usp15 | 7140 | -0.081 | 0.0381 | No |
| 75 | Dpp4 | 7147 | -0.082 | 0.0405 | No |
| 76 | Prkcd | 7299 | -0.094 | 0.0307 | No |
| 77 | Pik3ca | 7317 | -0.096 | 0.0326 | No |
| 78 | Dock10 | 7708 | -0.127 | 0.0032 | No |
| 79 | Irf2 | 7738 | -0.129 | 0.0052 | No |
| 80 | Lta4h | 7746 | -0.130 | 0.0092 | No |
| 81 | Stx4a | 7920 | -0.144 | -0.0008 | No |
| 82 | Pfn1 | 8251 | -0.170 | -0.0235 | No |
| 83 | Psen1 | 8398 | -0.182 | -0.0297 | No |
| 84 | Lcp2 | 8454 | -0.187 | -0.0279 | No |
| 85 | Pim1 | 8582 | -0.200 | -0.0319 | No |
| 86 | Gnb2 | 8677 | -0.209 | -0.0327 | No |
| 87 | Dgkh | 8753 | -0.215 | -0.0316 | No |
| 88 | Fcer1g | 8813 | -0.220 | -0.0290 | No |
| 89 | Casp9 | 8975 | -0.235 | -0.0347 | No |
| 90 | Gca | 9077 | -0.244 | -0.0348 | No |
| 91 | Gpd2 | 9159 | -0.253 | -0.0329 | No |
| 92 | Zeb1 | 9332 | -0.268 | -0.0384 | No |
| 93 | Fyn | 9422 | -0.277 | -0.0364 | No |
| 94 | Plscr1 | 9727 | -0.308 | -0.0519 | No |
| 95 | Ppp4c | 9737 | -0.309 | -0.0418 | No |
| 96 | Gnai2 | 9776 | -0.313 | -0.0341 | No |
| 97 | Ctsh | 9894 | -0.324 | -0.0328 | No |
| 98 | Pla2g4a | 10316 | -0.371 | -0.0563 | No |
| 99 | Ehd1 | 10622 | -0.411 | -0.0683 | No |
| 100 | Rhog | 10626 | -0.412 | -0.0540 | No |
| 101 | Lap3 | 10648 | -0.416 | -0.0412 | No |
| 102 | Akap10 | 10700 | -0.426 | -0.0305 | No |
| 103 | Dgkg | 11062 | -0.505 | -0.0441 | No |
| 104 | Casp7 | 11280 | -0.583 | -0.0424 | No |
| 105 | Casp3 | 11368 | -0.623 | -0.0279 | No |
| 106 | Calm1 | 11445 | -0.671 | -0.0108 | No |
| 107 | Rabif | 11488 | -0.708 | 0.0105 | No |