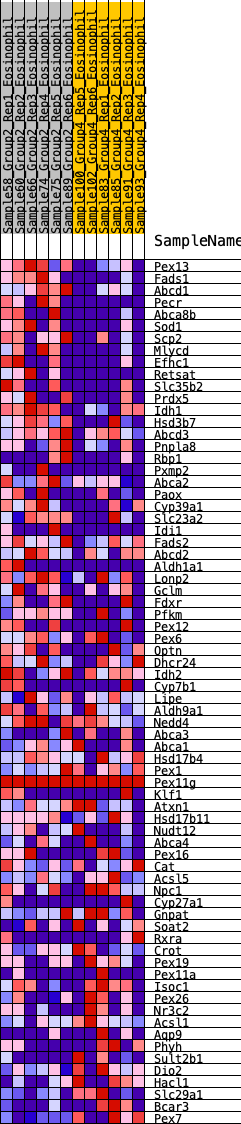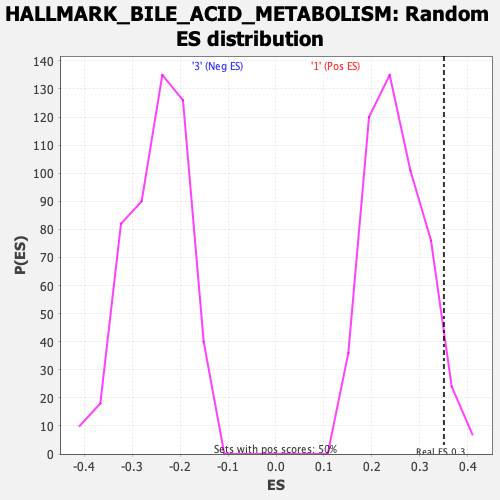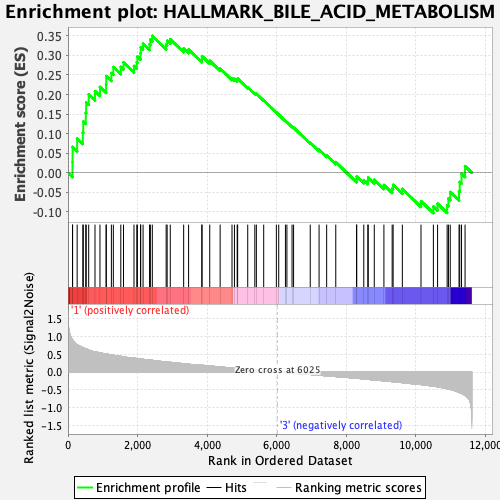
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.34991354 |
| Normalized Enrichment Score (NES) | 1.39342 |
| Nominal p-value | 0.0501002 |
| FDR q-value | 0.19490589 |
| FWER p-Value | 0.5 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex13 | 130 | 0.901 | 0.0274 | Yes |
| 2 | Fads1 | 132 | 0.901 | 0.0660 | Yes |
| 3 | Abcd1 | 263 | 0.760 | 0.0873 | Yes |
| 4 | Pecr | 427 | 0.687 | 0.1027 | Yes |
| 5 | Abca8b | 439 | 0.682 | 0.1310 | Yes |
| 6 | Sod1 | 511 | 0.652 | 0.1528 | Yes |
| 7 | Scp2 | 523 | 0.648 | 0.1796 | Yes |
| 8 | Mlycd | 595 | 0.619 | 0.2000 | Yes |
| 9 | Efhc1 | 776 | 0.564 | 0.2086 | Yes |
| 10 | Retsat | 919 | 0.537 | 0.2193 | Yes |
| 11 | Slc35b2 | 1096 | 0.504 | 0.2257 | Yes |
| 12 | Prdx5 | 1100 | 0.504 | 0.2471 | Yes |
| 13 | Idh1 | 1247 | 0.479 | 0.2550 | Yes |
| 14 | Hsd3b7 | 1303 | 0.470 | 0.2704 | Yes |
| 15 | Abcd3 | 1518 | 0.439 | 0.2707 | Yes |
| 16 | Pnpla8 | 1596 | 0.427 | 0.2823 | Yes |
| 17 | Rbp1 | 1897 | 0.389 | 0.2730 | Yes |
| 18 | Pxmp2 | 1978 | 0.379 | 0.2823 | Yes |
| 19 | Abca2 | 1995 | 0.378 | 0.2971 | Yes |
| 20 | Paox | 2085 | 0.367 | 0.3052 | Yes |
| 21 | Cyp39a1 | 2090 | 0.367 | 0.3206 | Yes |
| 22 | Slc23a2 | 2158 | 0.355 | 0.3300 | Yes |
| 23 | Idi1 | 2345 | 0.336 | 0.3283 | Yes |
| 24 | Fads2 | 2368 | 0.333 | 0.3407 | Yes |
| 25 | Abcd2 | 2424 | 0.327 | 0.3499 | Yes |
| 26 | Aldh1a1 | 2822 | 0.283 | 0.3276 | No |
| 27 | Lonp2 | 2850 | 0.282 | 0.3374 | No |
| 28 | Gclm | 2940 | 0.271 | 0.3413 | No |
| 29 | Fdxr | 3325 | 0.232 | 0.3180 | No |
| 30 | Pfkm | 3467 | 0.216 | 0.3150 | No |
| 31 | Pex12 | 3847 | 0.190 | 0.2903 | No |
| 32 | Pex6 | 3853 | 0.190 | 0.2981 | No |
| 33 | Optn | 4075 | 0.169 | 0.2862 | No |
| 34 | Dhcr24 | 4374 | 0.141 | 0.2664 | No |
| 35 | Idh2 | 4715 | 0.111 | 0.2417 | No |
| 36 | Cyp7b1 | 4785 | 0.104 | 0.2402 | No |
| 37 | Lipe | 4864 | 0.097 | 0.2376 | No |
| 38 | Aldh9a1 | 4877 | 0.096 | 0.2407 | No |
| 39 | Nedd4 | 5167 | 0.072 | 0.2187 | No |
| 40 | Abca3 | 5371 | 0.058 | 0.2036 | No |
| 41 | Abca1 | 5415 | 0.054 | 0.2022 | No |
| 42 | Hsd17b4 | 5626 | 0.033 | 0.1854 | No |
| 43 | Pex1 | 5990 | 0.003 | 0.1541 | No |
| 44 | Pex11g | 6058 | 0.000 | 0.1483 | No |
| 45 | Klf1 | 6251 | -0.013 | 0.1322 | No |
| 46 | Atxn1 | 6293 | -0.016 | 0.1293 | No |
| 47 | Hsd17b11 | 6441 | -0.029 | 0.1178 | No |
| 48 | Nudt12 | 6485 | -0.032 | 0.1155 | No |
| 49 | Abca4 | 6964 | -0.068 | 0.0770 | No |
| 50 | Pex16 | 7218 | -0.088 | 0.0588 | No |
| 51 | Cat | 7436 | -0.104 | 0.0445 | No |
| 52 | Acsl5 | 7698 | -0.126 | 0.0272 | No |
| 53 | Npc1 | 8296 | -0.175 | -0.0170 | No |
| 54 | Cyp27a1 | 8300 | -0.176 | -0.0097 | No |
| 55 | Gnpat | 8503 | -0.192 | -0.0190 | No |
| 56 | Soat2 | 8621 | -0.203 | -0.0204 | No |
| 57 | Rxra | 8630 | -0.205 | -0.0123 | No |
| 58 | Crot | 8805 | -0.219 | -0.0180 | No |
| 59 | Pex19 | 9082 | -0.245 | -0.0314 | No |
| 60 | Pex11a | 9319 | -0.267 | -0.0403 | No |
| 61 | Isoc1 | 9347 | -0.271 | -0.0310 | No |
| 62 | Pex26 | 9612 | -0.295 | -0.0412 | No |
| 63 | Nr3c2 | 10148 | -0.349 | -0.0726 | No |
| 64 | Acsl1 | 10504 | -0.394 | -0.0865 | No |
| 65 | Aqp9 | 10625 | -0.412 | -0.0792 | No |
| 66 | Phyh | 10900 | -0.465 | -0.0830 | No |
| 67 | Sult2b1 | 10937 | -0.473 | -0.0659 | No |
| 68 | Dio2 | 10989 | -0.484 | -0.0495 | No |
| 69 | Hacl1 | 11242 | -0.570 | -0.0469 | No |
| 70 | Slc29a1 | 11262 | -0.578 | -0.0237 | No |
| 71 | Bcar3 | 11313 | -0.597 | -0.0025 | No |
| 72 | Pex7 | 11415 | -0.653 | 0.0168 | No |