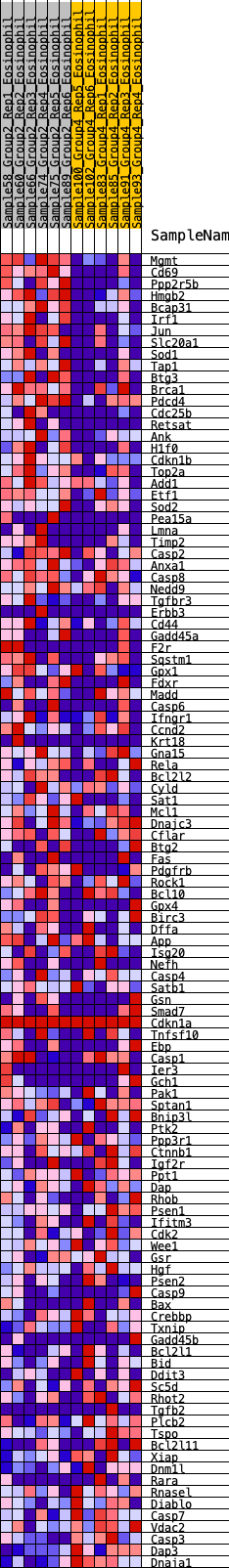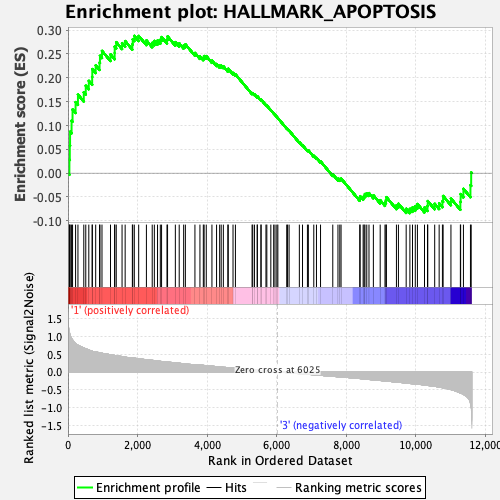
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.28802097 |
| Normalized Enrichment Score (NES) | 1.2636114 |
| Nominal p-value | 0.12138728 |
| FDR q-value | 0.30064923 |
| FWER p-Value | 0.771 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mgmt | 40 | 1.107 | 0.0283 | Yes |
| 2 | Cd69 | 52 | 1.072 | 0.0580 | Yes |
| 3 | Ppp2r5b | 60 | 1.033 | 0.0870 | Yes |
| 4 | Hmgb2 | 103 | 0.934 | 0.1102 | Yes |
| 5 | Bcap31 | 131 | 0.901 | 0.1336 | Yes |
| 6 | Irf1 | 218 | 0.798 | 0.1490 | Yes |
| 7 | Jun | 286 | 0.753 | 0.1648 | Yes |
| 8 | Slc20a1 | 454 | 0.676 | 0.1696 | Yes |
| 9 | Sod1 | 511 | 0.652 | 0.1834 | Yes |
| 10 | Tap1 | 598 | 0.618 | 0.1937 | Yes |
| 11 | Btg3 | 695 | 0.580 | 0.2020 | Yes |
| 12 | Brca1 | 699 | 0.580 | 0.2183 | Yes |
| 13 | Pdcd4 | 796 | 0.561 | 0.2261 | Yes |
| 14 | Cdc25b | 907 | 0.539 | 0.2319 | Yes |
| 15 | Retsat | 919 | 0.537 | 0.2464 | Yes |
| 16 | Ank | 977 | 0.526 | 0.2565 | Yes |
| 17 | H1f0 | 1222 | 0.483 | 0.2491 | Yes |
| 18 | Cdkn1b | 1341 | 0.465 | 0.2522 | Yes |
| 19 | Top2a | 1343 | 0.464 | 0.2654 | Yes |
| 20 | Add1 | 1387 | 0.456 | 0.2747 | Yes |
| 21 | Etf1 | 1554 | 0.433 | 0.2727 | Yes |
| 22 | Sod2 | 1643 | 0.419 | 0.2771 | Yes |
| 23 | Pea15a | 1850 | 0.392 | 0.2704 | Yes |
| 24 | Lmna | 1861 | 0.391 | 0.2807 | Yes |
| 25 | Timp2 | 1906 | 0.388 | 0.2880 | Yes |
| 26 | Casp2 | 2032 | 0.373 | 0.2879 | No |
| 27 | Anxa1 | 2254 | 0.345 | 0.2785 | No |
| 28 | Casp8 | 2421 | 0.327 | 0.2735 | No |
| 29 | Nedd9 | 2482 | 0.321 | 0.2774 | No |
| 30 | Tgfbr3 | 2574 | 0.308 | 0.2784 | No |
| 31 | Erbb3 | 2654 | 0.299 | 0.2800 | No |
| 32 | Cd44 | 2690 | 0.295 | 0.2855 | No |
| 33 | Gadd45a | 2846 | 0.282 | 0.2801 | No |
| 34 | F2r | 2862 | 0.281 | 0.2868 | No |
| 35 | Sqstm1 | 3086 | 0.259 | 0.2749 | No |
| 36 | Gpx1 | 3197 | 0.247 | 0.2724 | No |
| 37 | Fdxr | 3325 | 0.232 | 0.2680 | No |
| 38 | Madd | 3374 | 0.226 | 0.2703 | No |
| 39 | Casp6 | 3649 | 0.201 | 0.2522 | No |
| 40 | Ifngr1 | 3793 | 0.194 | 0.2453 | No |
| 41 | Ccnd2 | 3890 | 0.186 | 0.2423 | No |
| 42 | Krt18 | 3913 | 0.184 | 0.2457 | No |
| 43 | Gna15 | 3975 | 0.180 | 0.2456 | No |
| 44 | Rela | 4138 | 0.164 | 0.2362 | No |
| 45 | Bcl2l2 | 4271 | 0.152 | 0.2290 | No |
| 46 | Cyld | 4360 | 0.143 | 0.2255 | No |
| 47 | Sat1 | 4404 | 0.138 | 0.2257 | No |
| 48 | Mcl1 | 4466 | 0.131 | 0.2241 | No |
| 49 | Dnajc3 | 4596 | 0.123 | 0.2164 | No |
| 50 | Cflar | 4606 | 0.122 | 0.2191 | No |
| 51 | Btg2 | 4744 | 0.107 | 0.2103 | No |
| 52 | Fas | 4813 | 0.102 | 0.2073 | No |
| 53 | Pdgfrb | 5292 | 0.065 | 0.1676 | No |
| 54 | Rock1 | 5311 | 0.063 | 0.1679 | No |
| 55 | Bcl10 | 5360 | 0.059 | 0.1654 | No |
| 56 | Gpx4 | 5440 | 0.051 | 0.1600 | No |
| 57 | Birc3 | 5443 | 0.051 | 0.1612 | No |
| 58 | Dffa | 5548 | 0.042 | 0.1534 | No |
| 59 | App | 5550 | 0.041 | 0.1545 | No |
| 60 | Isg20 | 5691 | 0.028 | 0.1431 | No |
| 61 | Nefh | 5716 | 0.025 | 0.1417 | No |
| 62 | Casp4 | 5831 | 0.016 | 0.1322 | No |
| 63 | Satb1 | 5913 | 0.009 | 0.1255 | No |
| 64 | Gsn | 5951 | 0.006 | 0.1224 | No |
| 65 | Smad7 | 6004 | 0.002 | 0.1179 | No |
| 66 | Cdkn1a | 6033 | 0.000 | 0.1155 | No |
| 67 | Tnfsf10 | 6288 | -0.016 | 0.0939 | No |
| 68 | Ebp | 6312 | -0.017 | 0.0924 | No |
| 69 | Casp1 | 6356 | -0.021 | 0.0892 | No |
| 70 | Ier3 | 6651 | -0.046 | 0.0650 | No |
| 71 | Gch1 | 6742 | -0.050 | 0.0586 | No |
| 72 | Pak1 | 6883 | -0.062 | 0.0482 | No |
| 73 | Sptan1 | 6910 | -0.063 | 0.0478 | No |
| 74 | Bnip3l | 7066 | -0.074 | 0.0364 | No |
| 75 | Ptk2 | 7143 | -0.081 | 0.0321 | No |
| 76 | Ppp3r1 | 7260 | -0.091 | 0.0247 | No |
| 77 | Ctnnb1 | 7612 | -0.119 | -0.0025 | No |
| 78 | Igf2r | 7761 | -0.131 | -0.0116 | No |
| 79 | Ppt1 | 7814 | -0.135 | -0.0122 | No |
| 80 | Dap | 7850 | -0.138 | -0.0113 | No |
| 81 | Rhob | 8387 | -0.181 | -0.0527 | No |
| 82 | Psen1 | 8398 | -0.182 | -0.0484 | No |
| 83 | Ifitm3 | 8485 | -0.190 | -0.0504 | No |
| 84 | Cdk2 | 8513 | -0.193 | -0.0473 | No |
| 85 | Wee1 | 8537 | -0.196 | -0.0437 | No |
| 86 | Gsr | 8589 | -0.200 | -0.0424 | No |
| 87 | Hgf | 8653 | -0.207 | -0.0419 | No |
| 88 | Psen2 | 8779 | -0.217 | -0.0465 | No |
| 89 | Casp9 | 8975 | -0.235 | -0.0568 | No |
| 90 | Bax | 9113 | -0.248 | -0.0616 | No |
| 91 | Crebbp | 9140 | -0.252 | -0.0566 | No |
| 92 | Txnip | 9153 | -0.252 | -0.0504 | No |
| 93 | Gadd45b | 9442 | -0.279 | -0.0675 | No |
| 94 | Bcl2l1 | 9499 | -0.285 | -0.0642 | No |
| 95 | Bid | 9722 | -0.308 | -0.0747 | No |
| 96 | Ddit3 | 9829 | -0.319 | -0.0748 | No |
| 97 | Sc5d | 9904 | -0.326 | -0.0719 | No |
| 98 | Rhot2 | 9984 | -0.333 | -0.0692 | No |
| 99 | Tgfb2 | 10045 | -0.337 | -0.0648 | No |
| 100 | Plcb2 | 10249 | -0.363 | -0.0720 | No |
| 101 | Tspo | 10335 | -0.373 | -0.0687 | No |
| 102 | Bcl2l11 | 10343 | -0.375 | -0.0585 | No |
| 103 | Xiap | 10542 | -0.399 | -0.0643 | No |
| 104 | Dnm1l | 10670 | -0.421 | -0.0633 | No |
| 105 | Rara | 10764 | -0.436 | -0.0589 | No |
| 106 | Rnasel | 10784 | -0.442 | -0.0479 | No |
| 107 | Diablo | 11009 | -0.489 | -0.0533 | No |
| 108 | Casp7 | 11280 | -0.583 | -0.0601 | No |
| 109 | Vdac2 | 11288 | -0.587 | -0.0439 | No |
| 110 | Casp3 | 11368 | -0.623 | -0.0329 | No |
| 111 | Dap3 | 11570 | -0.878 | -0.0252 | No |
| 112 | Dnaja1 | 11591 | -0.994 | 0.0016 | No |