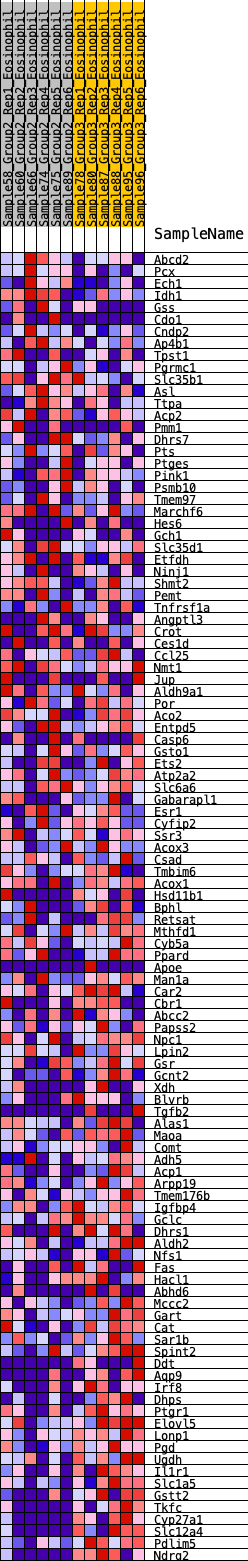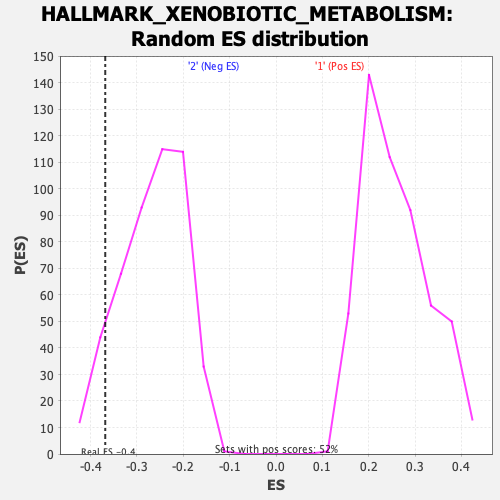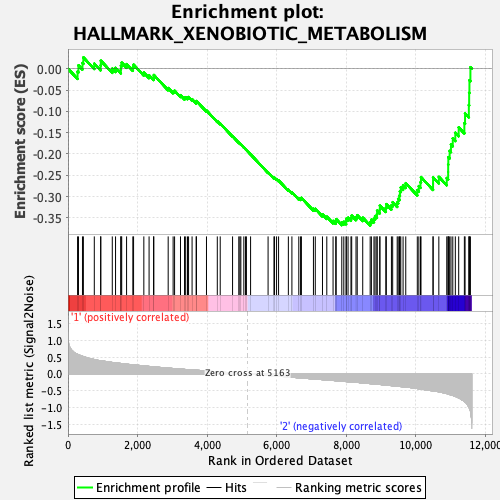
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.3682646 |
| Normalized Enrichment Score (NES) | -1.3861578 |
| Nominal p-value | 0.0875 |
| FDR q-value | 0.25807145 |
| FWER p-Value | 0.64 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcd2 | 275 | 0.586 | -0.0068 | No |
| 2 | Pcx | 301 | 0.575 | 0.0079 | No |
| 3 | Ech1 | 416 | 0.529 | 0.0135 | No |
| 4 | Idh1 | 442 | 0.519 | 0.0265 | No |
| 5 | Gss | 756 | 0.431 | 0.0119 | No |
| 6 | Cdo1 | 938 | 0.393 | 0.0077 | No |
| 7 | Cndp2 | 944 | 0.393 | 0.0188 | No |
| 8 | Ap4b1 | 1273 | 0.344 | 0.0003 | No |
| 9 | Tpst1 | 1367 | 0.331 | 0.0019 | No |
| 10 | Pgrmc1 | 1519 | 0.313 | -0.0020 | No |
| 11 | Slc35b1 | 1522 | 0.313 | 0.0069 | No |
| 12 | Asl | 1544 | 0.310 | 0.0142 | No |
| 13 | Ttpa | 1684 | 0.293 | 0.0107 | No |
| 14 | Acp2 | 1864 | 0.274 | 0.0032 | No |
| 15 | Pmm1 | 1885 | 0.272 | 0.0094 | No |
| 16 | Dhrs7 | 2180 | 0.242 | -0.0091 | No |
| 17 | Pts | 2331 | 0.225 | -0.0155 | No |
| 18 | Ptges | 2459 | 0.213 | -0.0203 | No |
| 19 | Pink1 | 2468 | 0.212 | -0.0148 | No |
| 20 | Psmb10 | 2880 | 0.176 | -0.0454 | No |
| 21 | Tmem97 | 3022 | 0.164 | -0.0529 | No |
| 22 | Marchf6 | 3064 | 0.159 | -0.0518 | No |
| 23 | Hes6 | 3234 | 0.146 | -0.0622 | No |
| 24 | Gch1 | 3350 | 0.136 | -0.0682 | No |
| 25 | Slc35d1 | 3383 | 0.134 | -0.0671 | No |
| 26 | Etfdh | 3435 | 0.130 | -0.0677 | No |
| 27 | Ninj1 | 3466 | 0.129 | -0.0665 | No |
| 28 | Shmt2 | 3567 | 0.121 | -0.0717 | No |
| 29 | Pemt | 3684 | 0.113 | -0.0785 | No |
| 30 | Tnfrsf1a | 3693 | 0.112 | -0.0759 | No |
| 31 | Angptl3 | 3981 | 0.090 | -0.0982 | No |
| 32 | Crot | 4292 | 0.066 | -0.1232 | No |
| 33 | Ces1d | 4376 | 0.058 | -0.1287 | No |
| 34 | Ccl25 | 4731 | 0.033 | -0.1586 | No |
| 35 | Nmt1 | 4910 | 0.018 | -0.1735 | No |
| 36 | Jup | 4941 | 0.015 | -0.1757 | No |
| 37 | Aldh9a1 | 4969 | 0.013 | -0.1776 | No |
| 38 | Por | 5055 | 0.007 | -0.1848 | No |
| 39 | Aco2 | 5104 | 0.003 | -0.1889 | No |
| 40 | Entpd5 | 5129 | 0.002 | -0.1909 | No |
| 41 | Casp6 | 5249 | -0.006 | -0.2011 | No |
| 42 | Gsto1 | 5752 | -0.045 | -0.2434 | No |
| 43 | Ets2 | 5921 | -0.056 | -0.2564 | No |
| 44 | Atp2a2 | 5938 | -0.058 | -0.2561 | No |
| 45 | Slc6a6 | 5996 | -0.062 | -0.2592 | No |
| 46 | Gabarapl1 | 6054 | -0.066 | -0.2623 | No |
| 47 | Esr1 | 6335 | -0.088 | -0.2840 | No |
| 48 | Cyfip2 | 6437 | -0.097 | -0.2900 | No |
| 49 | Ssr3 | 6635 | -0.113 | -0.3038 | No |
| 50 | Acox3 | 6686 | -0.117 | -0.3047 | No |
| 51 | Csad | 6710 | -0.120 | -0.3032 | No |
| 52 | Tmbim6 | 7056 | -0.148 | -0.3289 | No |
| 53 | Acox1 | 7109 | -0.152 | -0.3289 | No |
| 54 | Hsd11b1 | 7316 | -0.168 | -0.3419 | No |
| 55 | Bphl | 7440 | -0.178 | -0.3474 | No |
| 56 | Retsat | 7620 | -0.193 | -0.3573 | No |
| 57 | Mthfd1 | 7698 | -0.200 | -0.3582 | No |
| 58 | Cyb5a | 7709 | -0.201 | -0.3531 | No |
| 59 | Ppard | 7871 | -0.215 | -0.3609 | Yes |
| 60 | Apoe | 7929 | -0.221 | -0.3594 | Yes |
| 61 | Man1a | 7996 | -0.226 | -0.3585 | Yes |
| 62 | Car2 | 8000 | -0.226 | -0.3521 | Yes |
| 63 | Cbr1 | 8051 | -0.230 | -0.3498 | Yes |
| 64 | Abcc2 | 8135 | -0.237 | -0.3500 | Yes |
| 65 | Papss2 | 8156 | -0.238 | -0.3448 | Yes |
| 66 | Npc1 | 8280 | -0.247 | -0.3483 | Yes |
| 67 | Lpin2 | 8315 | -0.250 | -0.3439 | Yes |
| 68 | Gsr | 8473 | -0.267 | -0.3497 | Yes |
| 69 | Gcnt2 | 8687 | -0.287 | -0.3599 | Yes |
| 70 | Xdh | 8722 | -0.290 | -0.3543 | Yes |
| 71 | Blvrb | 8796 | -0.297 | -0.3520 | Yes |
| 72 | Tgfb2 | 8828 | -0.299 | -0.3459 | Yes |
| 73 | Alas1 | 8881 | -0.304 | -0.3415 | Yes |
| 74 | Maoa | 8887 | -0.305 | -0.3331 | Yes |
| 75 | Comt | 8964 | -0.312 | -0.3305 | Yes |
| 76 | Adh5 | 8966 | -0.312 | -0.3215 | Yes |
| 77 | Acp1 | 9137 | -0.330 | -0.3266 | Yes |
| 78 | Arpp19 | 9151 | -0.333 | -0.3180 | Yes |
| 79 | Tmem176b | 9298 | -0.346 | -0.3205 | Yes |
| 80 | Igfbp4 | 9334 | -0.350 | -0.3133 | Yes |
| 81 | Gclc | 9464 | -0.365 | -0.3139 | Yes |
| 82 | Dhrs1 | 9493 | -0.369 | -0.3055 | Yes |
| 83 | Aldh2 | 9530 | -0.372 | -0.2978 | Yes |
| 84 | Nfs1 | 9542 | -0.373 | -0.2878 | Yes |
| 85 | Fas | 9567 | -0.374 | -0.2789 | Yes |
| 86 | Hacl1 | 9633 | -0.380 | -0.2734 | Yes |
| 87 | Abhd6 | 9710 | -0.386 | -0.2687 | Yes |
| 88 | Mccc2 | 10042 | -0.433 | -0.2848 | Yes |
| 89 | Gart | 10086 | -0.441 | -0.2756 | Yes |
| 90 | Cat | 10134 | -0.448 | -0.2666 | Yes |
| 91 | Sar1b | 10148 | -0.451 | -0.2545 | Yes |
| 92 | Spint2 | 10495 | -0.503 | -0.2699 | Yes |
| 93 | Ddt | 10496 | -0.503 | -0.2551 | Yes |
| 94 | Aqp9 | 10661 | -0.530 | -0.2539 | Yes |
| 95 | Irf8 | 10889 | -0.583 | -0.2566 | Yes |
| 96 | Dhps | 10929 | -0.598 | -0.2424 | Yes |
| 97 | Ptgr1 | 10930 | -0.598 | -0.2249 | Yes |
| 98 | Elovl5 | 10934 | -0.600 | -0.2076 | Yes |
| 99 | Lonp1 | 10972 | -0.612 | -0.1929 | Yes |
| 100 | Pgd | 11010 | -0.623 | -0.1779 | Yes |
| 101 | Ugdh | 11062 | -0.640 | -0.1636 | Yes |
| 102 | Il1r1 | 11135 | -0.669 | -0.1502 | Yes |
| 103 | Slc1a5 | 11232 | -0.713 | -0.1377 | Yes |
| 104 | Gstt2 | 11395 | -0.819 | -0.1278 | Yes |
| 105 | Tkfc | 11415 | -0.836 | -0.1050 | Yes |
| 106 | Cyp27a1 | 11524 | -0.993 | -0.0853 | Yes |
| 107 | Slc12a4 | 11533 | -1.006 | -0.0565 | Yes |
| 108 | Pdlim5 | 11541 | -1.028 | -0.0270 | Yes |
| 109 | Ndrg2 | 11569 | -1.122 | 0.0035 | Yes |