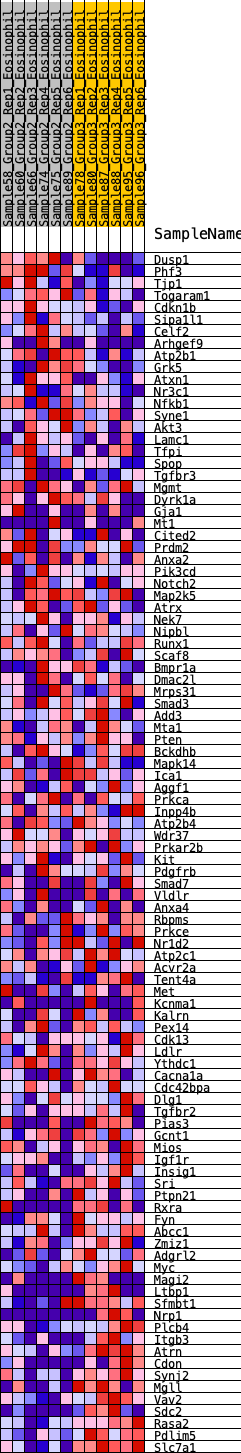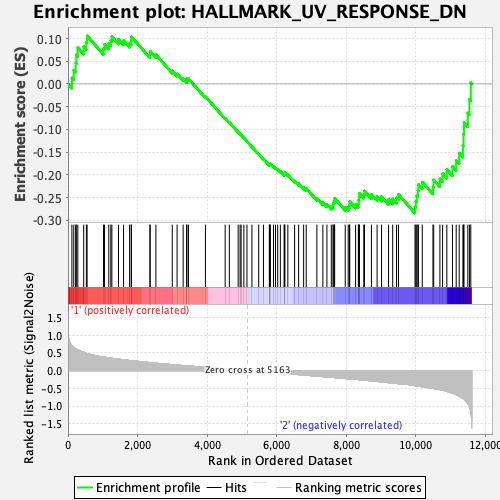
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | -0.2839651 |
| Normalized Enrichment Score (NES) | -1.2288036 |
| Nominal p-value | 0.172 |
| FDR q-value | 0.49243948 |
| FWER p-Value | 0.898 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp1 | 112 | 0.704 | 0.0135 | No |
| 2 | Phf3 | 166 | 0.651 | 0.0303 | No |
| 3 | Tjp1 | 220 | 0.610 | 0.0458 | No |
| 4 | Togaram1 | 239 | 0.603 | 0.0641 | No |
| 5 | Cdkn1b | 282 | 0.583 | 0.0797 | No |
| 6 | Sipa1l1 | 451 | 0.517 | 0.0821 | No |
| 7 | Celf2 | 528 | 0.489 | 0.0916 | No |
| 8 | Arhgef9 | 549 | 0.482 | 0.1058 | No |
| 9 | Atp2b1 | 1020 | 0.383 | 0.0775 | No |
| 10 | Grk5 | 1051 | 0.379 | 0.0874 | No |
| 11 | Atxn1 | 1173 | 0.357 | 0.0886 | No |
| 12 | Nr3c1 | 1231 | 0.350 | 0.0952 | No |
| 13 | Nfkb1 | 1261 | 0.345 | 0.1040 | No |
| 14 | Syne1 | 1452 | 0.321 | 0.0981 | No |
| 15 | Akt3 | 1597 | 0.302 | 0.0955 | No |
| 16 | Lamc1 | 1771 | 0.283 | 0.0898 | No |
| 17 | Tfpi | 1819 | 0.279 | 0.0949 | No |
| 18 | Spop | 1822 | 0.278 | 0.1039 | No |
| 19 | Tgfbr3 | 2355 | 0.223 | 0.0650 | No |
| 20 | Mgmt | 2361 | 0.222 | 0.0719 | No |
| 21 | Dyrk1a | 2527 | 0.206 | 0.0644 | No |
| 22 | Gja1 | 2997 | 0.165 | 0.0291 | No |
| 23 | Mt1 | 3136 | 0.154 | 0.0221 | No |
| 24 | Cited2 | 3310 | 0.140 | 0.0117 | No |
| 25 | Prdm2 | 3413 | 0.132 | 0.0072 | No |
| 26 | Anxa2 | 3416 | 0.132 | 0.0114 | No |
| 27 | Pik3cd | 3463 | 0.129 | 0.0116 | No |
| 28 | Notch2 | 3951 | 0.093 | -0.0276 | No |
| 29 | Map2k5 | 4518 | 0.049 | -0.0752 | No |
| 30 | Atrx | 4638 | 0.039 | -0.0842 | No |
| 31 | Nek7 | 4891 | 0.019 | -0.1055 | No |
| 32 | Nipbl | 4947 | 0.015 | -0.1098 | No |
| 33 | Runx1 | 4977 | 0.013 | -0.1119 | No |
| 34 | Scaf8 | 5056 | 0.007 | -0.1185 | No |
| 35 | Bmpr1a | 5145 | 0.001 | -0.1261 | No |
| 36 | Dmac2l | 5288 | -0.010 | -0.1381 | No |
| 37 | Mrps31 | 5482 | -0.024 | -0.1541 | No |
| 38 | Smad3 | 5619 | -0.035 | -0.1648 | No |
| 39 | Add3 | 5788 | -0.047 | -0.1778 | No |
| 40 | Mta1 | 5796 | -0.047 | -0.1769 | No |
| 41 | Pten | 5803 | -0.048 | -0.1758 | No |
| 42 | Bckdhb | 5817 | -0.049 | -0.1753 | No |
| 43 | Mapk14 | 5907 | -0.055 | -0.1813 | No |
| 44 | Ica1 | 5966 | -0.060 | -0.1843 | No |
| 45 | Aggf1 | 6028 | -0.064 | -0.1875 | No |
| 46 | Prkca | 6105 | -0.070 | -0.1918 | No |
| 47 | Inpp4b | 6215 | -0.078 | -0.1987 | No |
| 48 | Atp2b4 | 6220 | -0.078 | -0.1965 | No |
| 49 | Wdr37 | 6235 | -0.079 | -0.1951 | No |
| 50 | Prkar2b | 6320 | -0.087 | -0.1995 | No |
| 51 | Kit | 6515 | -0.104 | -0.2130 | No |
| 52 | Pdgfrb | 6630 | -0.112 | -0.2192 | No |
| 53 | Smad7 | 6774 | -0.125 | -0.2275 | No |
| 54 | Vldlr | 6848 | -0.131 | -0.2295 | No |
| 55 | Anxa4 | 7155 | -0.156 | -0.2510 | No |
| 56 | Rbpms | 7327 | -0.169 | -0.2603 | No |
| 57 | Prkce | 7443 | -0.178 | -0.2644 | No |
| 58 | Nr1d2 | 7570 | -0.188 | -0.2692 | No |
| 59 | Atp2c1 | 7622 | -0.193 | -0.2672 | No |
| 60 | Acvr2a | 7625 | -0.193 | -0.2610 | No |
| 61 | Tent4a | 7657 | -0.196 | -0.2573 | No |
| 62 | Met | 7666 | -0.197 | -0.2515 | No |
| 63 | Kcnma1 | 7970 | -0.224 | -0.2705 | Yes |
| 64 | Kalrn | 8055 | -0.230 | -0.2702 | Yes |
| 65 | Pex14 | 8089 | -0.233 | -0.2654 | Yes |
| 66 | Cdk13 | 8100 | -0.234 | -0.2586 | Yes |
| 67 | Ldlr | 8268 | -0.246 | -0.2650 | Yes |
| 68 | Ythdc1 | 8347 | -0.253 | -0.2634 | Yes |
| 69 | Cacna1a | 8353 | -0.253 | -0.2555 | Yes |
| 70 | Cdc42bpa | 8371 | -0.256 | -0.2486 | Yes |
| 71 | Dlg1 | 8376 | -0.257 | -0.2405 | Yes |
| 72 | Tgfbr2 | 8503 | -0.270 | -0.2425 | Yes |
| 73 | Pias3 | 8524 | -0.271 | -0.2353 | Yes |
| 74 | Gcnt1 | 8723 | -0.290 | -0.2430 | Yes |
| 75 | Mios | 8886 | -0.305 | -0.2470 | Yes |
| 76 | Igf1r | 9014 | -0.317 | -0.2476 | Yes |
| 77 | Insig1 | 9216 | -0.340 | -0.2539 | Yes |
| 78 | Sri | 9335 | -0.350 | -0.2526 | Yes |
| 79 | Ptpn21 | 9445 | -0.362 | -0.2502 | Yes |
| 80 | Rxra | 9500 | -0.369 | -0.2427 | Yes |
| 81 | Fyn | 9976 | -0.424 | -0.2700 | Yes |
| 82 | Abcc1 | 9999 | -0.427 | -0.2579 | Yes |
| 83 | Zmiz1 | 10026 | -0.432 | -0.2459 | Yes |
| 84 | Adgrl2 | 10059 | -0.437 | -0.2343 | Yes |
| 85 | Myc | 10073 | -0.438 | -0.2210 | Yes |
| 86 | Magi2 | 10185 | -0.456 | -0.2156 | Yes |
| 87 | Ltbp1 | 10492 | -0.503 | -0.2257 | Yes |
| 88 | Sfmbt1 | 10514 | -0.505 | -0.2108 | Yes |
| 89 | Nrp1 | 10692 | -0.537 | -0.2085 | Yes |
| 90 | Plcb4 | 10769 | -0.554 | -0.1969 | Yes |
| 91 | Itgb3 | 10891 | -0.583 | -0.1882 | Yes |
| 92 | Atrn | 11053 | -0.636 | -0.1813 | Yes |
| 93 | Cdon | 11160 | -0.677 | -0.1682 | Yes |
| 94 | Synj2 | 11246 | -0.721 | -0.1518 | Yes |
| 95 | Mgll | 11353 | -0.785 | -0.1352 | Yes |
| 96 | Vav2 | 11371 | -0.797 | -0.1104 | Yes |
| 97 | Sdc2 | 11384 | -0.811 | -0.0848 | Yes |
| 98 | Rasa2 | 11494 | -0.931 | -0.0636 | Yes |
| 99 | Pdlim5 | 11541 | -1.028 | -0.0337 | Yes |
| 100 | Slc7a1 | 11580 | -1.201 | 0.0025 | Yes |