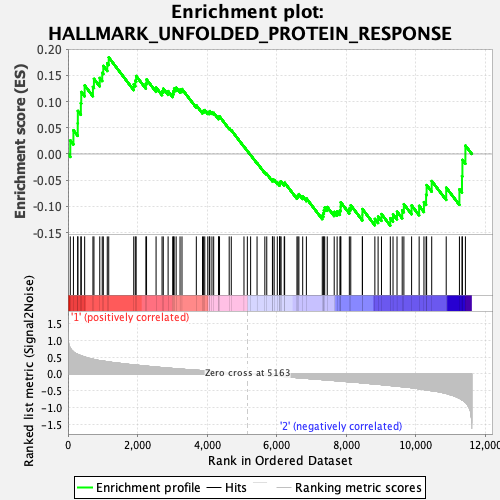
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.18400791 |
| Normalized Enrichment Score (NES) | 0.79525346 |
| Nominal p-value | 0.81060606 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Serp1 | 66 | 0.773 | 0.0259 | Yes |
| 2 | Mtrex | 156 | 0.661 | 0.0452 | Yes |
| 3 | Cebpb | 280 | 0.583 | 0.0584 | Yes |
| 4 | Bag3 | 283 | 0.583 | 0.0821 | Yes |
| 5 | Nfyb | 370 | 0.545 | 0.0969 | Yes |
| 6 | Nabp1 | 381 | 0.540 | 0.1182 | Yes |
| 7 | Eif4g1 | 479 | 0.508 | 0.1305 | Yes |
| 8 | Eif2s1 | 717 | 0.439 | 0.1279 | Yes |
| 9 | Herpud1 | 746 | 0.433 | 0.1432 | Yes |
| 10 | Hspa9 | 913 | 0.396 | 0.1450 | Yes |
| 11 | Arfgap1 | 984 | 0.388 | 0.1547 | Yes |
| 12 | Atf6 | 1015 | 0.383 | 0.1678 | Yes |
| 13 | Hsp90b1 | 1129 | 0.365 | 0.1729 | Yes |
| 14 | Nop14 | 1171 | 0.357 | 0.1840 | Yes |
| 15 | Kif5b | 1890 | 0.272 | 0.1328 | No |
| 16 | Tspyl2 | 1935 | 0.266 | 0.1398 | No |
| 17 | Banf1 | 1959 | 0.264 | 0.1487 | No |
| 18 | Hspa5 | 2239 | 0.236 | 0.1341 | No |
| 19 | Vegfa | 2258 | 0.234 | 0.1421 | No |
| 20 | Cks1b | 2532 | 0.206 | 0.1268 | No |
| 21 | Cebpg | 2704 | 0.192 | 0.1198 | No |
| 22 | Wipi1 | 2739 | 0.189 | 0.1246 | No |
| 23 | Preb | 2878 | 0.176 | 0.1198 | No |
| 24 | Sec11a | 3010 | 0.164 | 0.1151 | No |
| 25 | Tatdn2 | 3033 | 0.162 | 0.1198 | No |
| 26 | Dcp2 | 3052 | 0.160 | 0.1248 | No |
| 27 | Cnot6 | 3109 | 0.156 | 0.1263 | No |
| 28 | Atp6v0d1 | 3221 | 0.147 | 0.1227 | No |
| 29 | Ywhaz | 3277 | 0.143 | 0.1238 | No |
| 30 | Cnot2 | 3689 | 0.112 | 0.0927 | No |
| 31 | Exoc2 | 3867 | 0.099 | 0.0813 | No |
| 32 | Sec31a | 3898 | 0.097 | 0.0827 | No |
| 33 | Dnajb9 | 3932 | 0.094 | 0.0837 | No |
| 34 | Pop4 | 4018 | 0.087 | 0.0798 | No |
| 35 | Atf4 | 4063 | 0.084 | 0.0794 | No |
| 36 | Dcp1a | 4077 | 0.083 | 0.0817 | No |
| 37 | Gosr2 | 4138 | 0.078 | 0.0796 | No |
| 38 | Ddx10 | 4188 | 0.074 | 0.0784 | No |
| 39 | Shc1 | 4328 | 0.062 | 0.0689 | No |
| 40 | Slc30a5 | 4333 | 0.062 | 0.0710 | No |
| 41 | Ero1a | 4356 | 0.059 | 0.0715 | No |
| 42 | Khsrp | 4634 | 0.039 | 0.0491 | No |
| 43 | Srprb | 4694 | 0.036 | 0.0454 | No |
| 44 | Dnajc3 | 5059 | 0.007 | 0.0141 | No |
| 45 | Calr | 5156 | 0.000 | 0.0058 | No |
| 46 | Ssr1 | 5250 | -0.006 | -0.0020 | No |
| 47 | Rrp9 | 5435 | -0.021 | -0.0172 | No |
| 48 | Exosc4 | 5659 | -0.037 | -0.0350 | No |
| 49 | Dctn1 | 5714 | -0.042 | -0.0380 | No |
| 50 | Srpr | 5877 | -0.053 | -0.0499 | No |
| 51 | H2ax | 5885 | -0.054 | -0.0483 | No |
| 52 | Edem1 | 5927 | -0.057 | -0.0495 | No |
| 53 | Eif4ebp1 | 6017 | -0.063 | -0.0547 | No |
| 54 | Eif2ak3 | 6084 | -0.068 | -0.0576 | No |
| 55 | Spcs1 | 6085 | -0.068 | -0.0548 | No |
| 56 | Npm1 | 6097 | -0.069 | -0.0529 | No |
| 57 | Hyou1 | 6126 | -0.072 | -0.0524 | No |
| 58 | Eif4a3 | 6216 | -0.078 | -0.0570 | No |
| 59 | Pdia6 | 6228 | -0.079 | -0.0547 | No |
| 60 | Paip1 | 6582 | -0.109 | -0.0809 | No |
| 61 | Exosc10 | 6610 | -0.111 | -0.0787 | No |
| 62 | Imp3 | 6638 | -0.113 | -0.0764 | No |
| 63 | Cxxc1 | 6747 | -0.123 | -0.0808 | No |
| 64 | Eef2 | 6853 | -0.131 | -0.0845 | No |
| 65 | Mthfd2 | 7310 | -0.168 | -0.1173 | No |
| 66 | Eif4a1 | 7340 | -0.170 | -0.1129 | No |
| 67 | Exosc5 | 7357 | -0.172 | -0.1072 | No |
| 68 | Xbp1 | 7380 | -0.173 | -0.1021 | No |
| 69 | Xpot | 7453 | -0.179 | -0.1010 | No |
| 70 | Lsm4 | 7653 | -0.195 | -0.1103 | No |
| 71 | Parn | 7737 | -0.202 | -0.1092 | No |
| 72 | Zbtb17 | 7817 | -0.210 | -0.1075 | No |
| 73 | Eif4a2 | 7839 | -0.212 | -0.1006 | No |
| 74 | Rps14 | 7841 | -0.212 | -0.0920 | No |
| 75 | Lsm1 | 8088 | -0.233 | -0.1039 | No |
| 76 | Sdad1 | 8129 | -0.236 | -0.0977 | No |
| 77 | Edc4 | 8463 | -0.266 | -0.1157 | No |
| 78 | Pdia5 | 8464 | -0.266 | -0.1049 | No |
| 79 | Dnaja4 | 8822 | -0.299 | -0.1236 | No |
| 80 | Ern1 | 8915 | -0.307 | -0.1191 | No |
| 81 | Nhp2 | 9012 | -0.317 | -0.1145 | No |
| 82 | Aldh18a1 | 9266 | -0.344 | -0.1224 | No |
| 83 | Asns | 9344 | -0.350 | -0.1147 | No |
| 84 | Exosc2 | 9459 | -0.364 | -0.1098 | No |
| 85 | Wfs1 | 9608 | -0.378 | -0.1072 | No |
| 86 | Eif4e | 9656 | -0.381 | -0.0956 | No |
| 87 | Slc7a5 | 9878 | -0.410 | -0.0981 | No |
| 88 | Cnot4 | 10095 | -0.442 | -0.0988 | No |
| 89 | Fus | 10233 | -0.464 | -0.0917 | No |
| 90 | Nfya | 10295 | -0.475 | -0.0776 | No |
| 91 | Nolc1 | 10310 | -0.477 | -0.0592 | No |
| 92 | Exosc9 | 10456 | -0.497 | -0.0515 | No |
| 93 | Psat1 | 10873 | -0.576 | -0.0640 | No |
| 94 | Exosc1 | 11253 | -0.725 | -0.0673 | No |
| 95 | Yif1a | 11327 | -0.772 | -0.0420 | No |
| 96 | Nop56 | 11339 | -0.776 | -0.0112 | No |
| 97 | Dkc1 | 11425 | -0.846 | 0.0160 | No |

