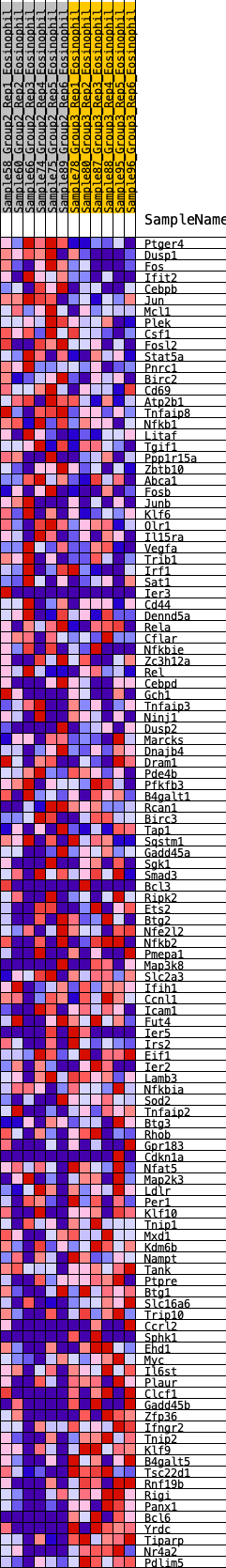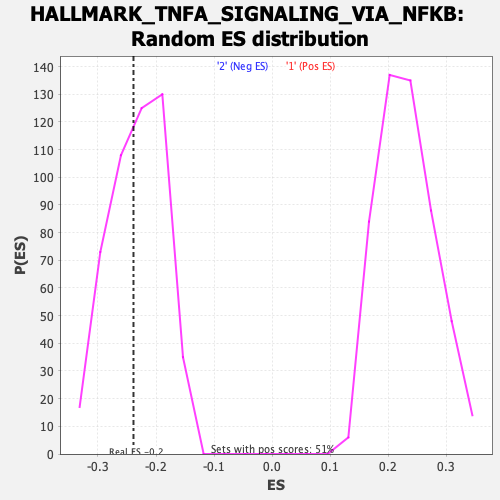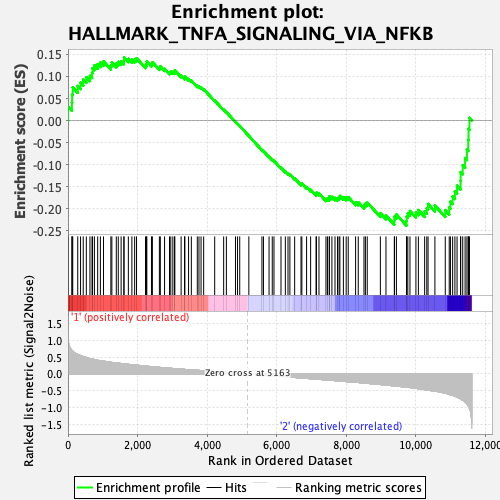
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | -0.23880422 |
| Normalized Enrichment Score (NES) | -1.0283463 |
| Nominal p-value | 0.42213115 |
| FDR q-value | 0.76878655 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ptger4 | 4 | 1.207 | 0.0316 | No |
| 2 | Dusp1 | 112 | 0.704 | 0.0409 | No |
| 3 | Fos | 119 | 0.695 | 0.0588 | No |
| 4 | Ifit2 | 136 | 0.680 | 0.0754 | No |
| 5 | Cebpb | 280 | 0.583 | 0.0784 | No |
| 6 | Jun | 361 | 0.548 | 0.0859 | No |
| 7 | Mcl1 | 438 | 0.520 | 0.0931 | No |
| 8 | Plek | 527 | 0.489 | 0.0984 | No |
| 9 | Csf1 | 631 | 0.458 | 0.1015 | No |
| 10 | Fosl2 | 691 | 0.444 | 0.1081 | No |
| 11 | Stat5a | 705 | 0.441 | 0.1187 | No |
| 12 | Pnrc1 | 759 | 0.431 | 0.1254 | No |
| 13 | Birc2 | 853 | 0.407 | 0.1281 | No |
| 14 | Cd69 | 931 | 0.394 | 0.1318 | No |
| 15 | Atp2b1 | 1020 | 0.383 | 0.1343 | No |
| 16 | Tnfaip8 | 1232 | 0.350 | 0.1252 | No |
| 17 | Nfkb1 | 1261 | 0.345 | 0.1319 | No |
| 18 | Litaf | 1388 | 0.329 | 0.1296 | No |
| 19 | Tgif1 | 1445 | 0.323 | 0.1333 | No |
| 20 | Ppp1r15a | 1527 | 0.312 | 0.1345 | No |
| 21 | Zbtb10 | 1606 | 0.301 | 0.1357 | No |
| 22 | Abca1 | 1611 | 0.301 | 0.1433 | No |
| 23 | Fosb | 1735 | 0.287 | 0.1402 | No |
| 24 | Junb | 1837 | 0.277 | 0.1387 | No |
| 25 | Klf6 | 1916 | 0.269 | 0.1391 | No |
| 26 | Olr1 | 1970 | 0.263 | 0.1414 | No |
| 27 | Il15ra | 2228 | 0.237 | 0.1253 | No |
| 28 | Vegfa | 2258 | 0.234 | 0.1290 | No |
| 29 | Trib1 | 2265 | 0.233 | 0.1346 | No |
| 30 | Irf1 | 2400 | 0.219 | 0.1288 | No |
| 31 | Sat1 | 2426 | 0.216 | 0.1323 | No |
| 32 | Ier3 | 2627 | 0.197 | 0.1201 | No |
| 33 | Cd44 | 2654 | 0.195 | 0.1230 | No |
| 34 | Dennd5a | 2776 | 0.185 | 0.1174 | No |
| 35 | Rela | 2918 | 0.173 | 0.1097 | No |
| 36 | Cflar | 2955 | 0.169 | 0.1110 | No |
| 37 | Nfkbie | 3014 | 0.164 | 0.1103 | No |
| 38 | Zc3h12a | 3065 | 0.159 | 0.1102 | No |
| 39 | Rel | 3069 | 0.159 | 0.1141 | No |
| 40 | Cebpd | 3252 | 0.144 | 0.1021 | No |
| 41 | Gch1 | 3350 | 0.136 | 0.0973 | No |
| 42 | Tnfaip3 | 3360 | 0.136 | 0.1001 | No |
| 43 | Ninj1 | 3466 | 0.129 | 0.0943 | No |
| 44 | Dusp2 | 3546 | 0.122 | 0.0907 | No |
| 45 | Marcks | 3717 | 0.110 | 0.0788 | No |
| 46 | Dnajb4 | 3765 | 0.107 | 0.0776 | No |
| 47 | Dram1 | 3825 | 0.102 | 0.0751 | No |
| 48 | Pde4b | 3900 | 0.097 | 0.0712 | No |
| 49 | Pfkfb3 | 4217 | 0.072 | 0.0456 | No |
| 50 | B4galt1 | 4475 | 0.052 | 0.0246 | No |
| 51 | Rcan1 | 4553 | 0.045 | 0.0191 | No |
| 52 | Birc3 | 4815 | 0.026 | -0.0029 | No |
| 53 | Tap1 | 4874 | 0.020 | -0.0074 | No |
| 54 | Sqstm1 | 4937 | 0.016 | -0.0124 | No |
| 55 | Gadd45a | 5199 | -0.002 | -0.0351 | No |
| 56 | Sgk1 | 5575 | -0.031 | -0.0669 | No |
| 57 | Smad3 | 5619 | -0.035 | -0.0697 | No |
| 58 | Bcl3 | 5780 | -0.047 | -0.0824 | No |
| 59 | Ripk2 | 5872 | -0.053 | -0.0889 | No |
| 60 | Ets2 | 5921 | -0.056 | -0.0916 | No |
| 61 | Btg2 | 6123 | -0.072 | -0.1072 | No |
| 62 | Nfe2l2 | 6248 | -0.080 | -0.1159 | No |
| 63 | Nfkb2 | 6325 | -0.088 | -0.1201 | No |
| 64 | Pmepa1 | 6379 | -0.092 | -0.1223 | No |
| 65 | Map3k8 | 6516 | -0.104 | -0.1314 | No |
| 66 | Slc2a3 | 6699 | -0.119 | -0.1441 | No |
| 67 | Ifih1 | 6723 | -0.121 | -0.1429 | No |
| 68 | Ccnl1 | 6859 | -0.132 | -0.1512 | No |
| 69 | Icam1 | 6975 | -0.141 | -0.1574 | No |
| 70 | Fut4 | 7131 | -0.153 | -0.1669 | No |
| 71 | Ier5 | 7143 | -0.154 | -0.1637 | No |
| 72 | Irs2 | 7215 | -0.159 | -0.1657 | No |
| 73 | Eif1 | 7416 | -0.176 | -0.1784 | No |
| 74 | Ier2 | 7459 | -0.179 | -0.1773 | No |
| 75 | Lamb3 | 7511 | -0.183 | -0.1769 | No |
| 76 | Nfkbia | 7514 | -0.184 | -0.1722 | No |
| 77 | Sod2 | 7587 | -0.190 | -0.1735 | No |
| 78 | Tnfaip2 | 7676 | -0.197 | -0.1759 | No |
| 79 | Btg3 | 7748 | -0.204 | -0.1767 | No |
| 80 | Rhob | 7792 | -0.207 | -0.1750 | No |
| 81 | Gpr183 | 7816 | -0.210 | -0.1714 | No |
| 82 | Cdkn1a | 7917 | -0.220 | -0.1743 | No |
| 83 | Nfat5 | 7997 | -0.226 | -0.1752 | No |
| 84 | Map2k3 | 8056 | -0.230 | -0.1741 | No |
| 85 | Ldlr | 8268 | -0.246 | -0.1860 | No |
| 86 | Per1 | 8344 | -0.253 | -0.1858 | No |
| 87 | Klf10 | 8509 | -0.270 | -0.1929 | No |
| 88 | Tnip1 | 8550 | -0.273 | -0.1892 | No |
| 89 | Mxd1 | 8603 | -0.278 | -0.1864 | No |
| 90 | Kdm6b | 8980 | -0.313 | -0.2108 | No |
| 91 | Nampt | 9141 | -0.331 | -0.2160 | No |
| 92 | Tank | 9382 | -0.353 | -0.2275 | No |
| 93 | Ptpre | 9385 | -0.353 | -0.2184 | No |
| 94 | Btg1 | 9441 | -0.362 | -0.2136 | No |
| 95 | Slc16a6 | 9732 | -0.389 | -0.2285 | Yes |
| 96 | Trip10 | 9737 | -0.389 | -0.2186 | Yes |
| 97 | Ccrl2 | 9769 | -0.392 | -0.2109 | Yes |
| 98 | Sphk1 | 9831 | -0.402 | -0.2056 | Yes |
| 99 | Ehd1 | 10002 | -0.428 | -0.2090 | Yes |
| 100 | Myc | 10073 | -0.438 | -0.2035 | Yes |
| 101 | Il6st | 10253 | -0.468 | -0.2067 | Yes |
| 102 | Plaur | 10315 | -0.478 | -0.1994 | Yes |
| 103 | Clcf1 | 10351 | -0.483 | -0.1896 | Yes |
| 104 | Gadd45b | 10548 | -0.510 | -0.1932 | Yes |
| 105 | Zfp36 | 10846 | -0.570 | -0.2040 | Yes |
| 106 | Ifngr2 | 10960 | -0.608 | -0.1977 | Yes |
| 107 | Tnip2 | 10994 | -0.618 | -0.1842 | Yes |
| 108 | Klf9 | 11058 | -0.637 | -0.1728 | Yes |
| 109 | B4galt5 | 11123 | -0.662 | -0.1609 | Yes |
| 110 | Tsc22d1 | 11185 | -0.691 | -0.1479 | Yes |
| 111 | Rnf19b | 11286 | -0.746 | -0.1369 | Yes |
| 112 | Rigi | 11289 | -0.747 | -0.1173 | Yes |
| 113 | Panx1 | 11348 | -0.780 | -0.1017 | Yes |
| 114 | Bcl6 | 11416 | -0.837 | -0.0853 | Yes |
| 115 | Yrdc | 11469 | -0.894 | -0.0662 | Yes |
| 116 | Tiparp | 11507 | -0.956 | -0.0441 | Yes |
| 117 | Nr4a2 | 11515 | -0.968 | -0.0191 | Yes |
| 118 | Pdlim5 | 11541 | -1.028 | 0.0059 | Yes |