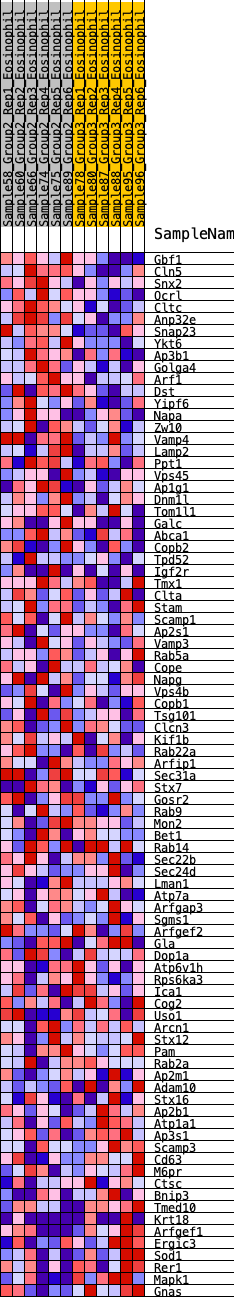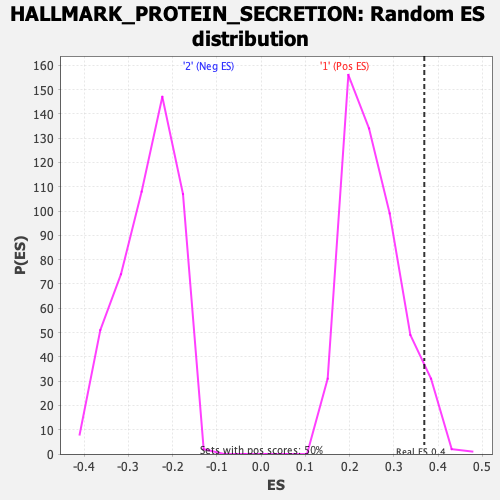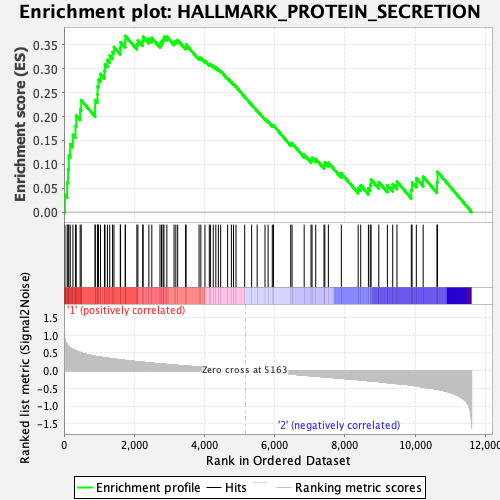
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.36878335 |
| Normalized Enrichment Score (NES) | 1.4675076 |
| Nominal p-value | 0.06560636 |
| FDR q-value | 0.47081757 |
| FWER p-Value | 0.448 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gbf1 | 26 | 0.884 | 0.0361 | Yes |
| 2 | Cln5 | 90 | 0.729 | 0.0622 | Yes |
| 3 | Snx2 | 116 | 0.700 | 0.0904 | Yes |
| 4 | Ocrl | 137 | 0.680 | 0.1181 | Yes |
| 5 | Cltc | 178 | 0.639 | 0.1424 | Yes |
| 6 | Anp32e | 254 | 0.596 | 0.1617 | Yes |
| 7 | Snap23 | 326 | 0.564 | 0.1800 | Yes |
| 8 | Ykt6 | 347 | 0.554 | 0.2023 | Yes |
| 9 | Ap3b1 | 460 | 0.512 | 0.2148 | Yes |
| 10 | Golga4 | 491 | 0.504 | 0.2341 | Yes |
| 11 | Arf1 | 886 | 0.402 | 0.2173 | Yes |
| 12 | Dst | 888 | 0.401 | 0.2346 | Yes |
| 13 | Yipf6 | 953 | 0.392 | 0.2460 | Yes |
| 14 | Napa | 956 | 0.391 | 0.2628 | Yes |
| 15 | Zw10 | 983 | 0.388 | 0.2774 | Yes |
| 16 | Vamp4 | 1039 | 0.381 | 0.2891 | Yes |
| 17 | Lamp2 | 1152 | 0.361 | 0.2950 | Yes |
| 18 | Ppt1 | 1168 | 0.358 | 0.3093 | Yes |
| 19 | Vps45 | 1237 | 0.349 | 0.3185 | Yes |
| 20 | Ap1g1 | 1303 | 0.339 | 0.3275 | Yes |
| 21 | Dnm1l | 1383 | 0.329 | 0.3349 | Yes |
| 22 | Tom1l1 | 1423 | 0.325 | 0.3457 | Yes |
| 23 | Galc | 1603 | 0.302 | 0.3432 | Yes |
| 24 | Abca1 | 1611 | 0.301 | 0.3556 | Yes |
| 25 | Copb2 | 1745 | 0.286 | 0.3565 | Yes |
| 26 | Tpd52 | 1747 | 0.285 | 0.3688 | Yes |
| 27 | Igf2r | 2076 | 0.253 | 0.3513 | No |
| 28 | Tmx1 | 2104 | 0.250 | 0.3598 | No |
| 29 | Clta | 2242 | 0.235 | 0.3581 | No |
| 30 | Stam | 2253 | 0.234 | 0.3674 | No |
| 31 | Scamp1 | 2417 | 0.217 | 0.3626 | No |
| 32 | Ap2s1 | 2501 | 0.209 | 0.3645 | No |
| 33 | Vamp3 | 2734 | 0.190 | 0.3526 | No |
| 34 | Rab5a | 2778 | 0.185 | 0.3569 | No |
| 35 | Cope | 2821 | 0.182 | 0.3611 | No |
| 36 | Napg | 2847 | 0.179 | 0.3667 | No |
| 37 | Vps4b | 2929 | 0.172 | 0.3672 | No |
| 38 | Copb1 | 3134 | 0.154 | 0.3561 | No |
| 39 | Tsg101 | 3185 | 0.150 | 0.3583 | No |
| 40 | Clcn3 | 3237 | 0.146 | 0.3602 | No |
| 41 | Kif1b | 3462 | 0.129 | 0.3463 | No |
| 42 | Rab22a | 3477 | 0.128 | 0.3507 | No |
| 43 | Arfip1 | 3846 | 0.100 | 0.3231 | No |
| 44 | Sec31a | 3898 | 0.097 | 0.3229 | No |
| 45 | Stx7 | 4014 | 0.087 | 0.3167 | No |
| 46 | Gosr2 | 4138 | 0.078 | 0.3093 | No |
| 47 | Rab9 | 4170 | 0.074 | 0.3099 | No |
| 48 | Mon2 | 4250 | 0.069 | 0.3060 | No |
| 49 | Bet1 | 4324 | 0.062 | 0.3024 | No |
| 50 | Rab14 | 4396 | 0.057 | 0.2987 | No |
| 51 | Sec22b | 4462 | 0.053 | 0.2953 | No |
| 52 | Sec24d | 4660 | 0.038 | 0.2799 | No |
| 53 | Lman1 | 4767 | 0.030 | 0.2720 | No |
| 54 | Atp7a | 4827 | 0.025 | 0.2679 | No |
| 55 | Arfgap3 | 4898 | 0.018 | 0.2626 | No |
| 56 | Sgms1 | 5146 | 0.001 | 0.2412 | No |
| 57 | Arfgef2 | 5344 | -0.013 | 0.2247 | No |
| 58 | Gla | 5501 | -0.026 | 0.2123 | No |
| 59 | Dop1a | 5725 | -0.043 | 0.1948 | No |
| 60 | Atp6v1h | 5813 | -0.048 | 0.1893 | No |
| 61 | Rps6ka3 | 5932 | -0.057 | 0.1816 | No |
| 62 | Ica1 | 5966 | -0.060 | 0.1813 | No |
| 63 | Cog2 | 6457 | -0.099 | 0.1431 | No |
| 64 | Uso1 | 6495 | -0.102 | 0.1443 | No |
| 65 | Arcn1 | 6840 | -0.130 | 0.1201 | No |
| 66 | Stx12 | 7035 | -0.145 | 0.1096 | No |
| 67 | Pam | 7064 | -0.148 | 0.1136 | No |
| 68 | Rab2a | 7169 | -0.156 | 0.1113 | No |
| 69 | Ap2m1 | 7405 | -0.175 | 0.0985 | No |
| 70 | Adam10 | 7429 | -0.177 | 0.1042 | No |
| 71 | Stx16 | 7529 | -0.185 | 0.1036 | No |
| 72 | Ap2b1 | 7898 | -0.217 | 0.0811 | No |
| 73 | Atp1a1 | 8378 | -0.257 | 0.0507 | No |
| 74 | Ap3s1 | 8450 | -0.265 | 0.0560 | No |
| 75 | Scamp3 | 8670 | -0.285 | 0.0493 | No |
| 76 | Cd63 | 8721 | -0.290 | 0.0576 | No |
| 77 | M6pr | 8746 | -0.292 | 0.0681 | No |
| 78 | Ctsc | 8965 | -0.312 | 0.0627 | No |
| 79 | Bnip3 | 9210 | -0.339 | 0.0563 | No |
| 80 | Tmed10 | 9362 | -0.351 | 0.0584 | No |
| 81 | Krt18 | 9483 | -0.367 | 0.0639 | No |
| 82 | Arfgef1 | 9891 | -0.411 | 0.0464 | No |
| 83 | Ergic3 | 9913 | -0.414 | 0.0626 | No |
| 84 | Sod1 | 10034 | -0.432 | 0.0709 | No |
| 85 | Rer1 | 10229 | -0.464 | 0.0742 | No |
| 86 | Mapk1 | 10622 | -0.522 | 0.0628 | No |
| 87 | Gnas | 10638 | -0.526 | 0.0843 | No |