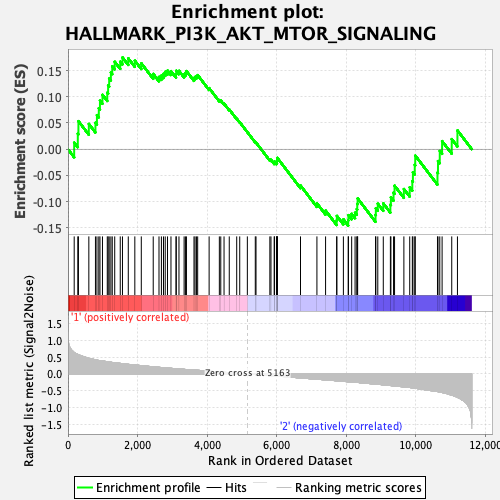
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.17502144 |
| Normalized Enrichment Score (NES) | 0.78021127 |
| Nominal p-value | 0.8871287 |
| FDR q-value | 0.9621771 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 178 | 0.639 | 0.0127 | Yes |
| 2 | Cdkn1b | 282 | 0.583 | 0.0296 | Yes |
| 3 | Map2k6 | 300 | 0.576 | 0.0535 | Yes |
| 4 | Ptpn11 | 599 | 0.468 | 0.0483 | Yes |
| 5 | Plcb1 | 789 | 0.422 | 0.0505 | Yes |
| 6 | Ralb | 832 | 0.412 | 0.0650 | Yes |
| 7 | Arf1 | 886 | 0.402 | 0.0782 | Yes |
| 8 | Akt1 | 919 | 0.395 | 0.0928 | Yes |
| 9 | Actr3 | 991 | 0.387 | 0.1037 | Yes |
| 10 | Hsp90b1 | 1129 | 0.365 | 0.1080 | Yes |
| 11 | E2f1 | 1154 | 0.361 | 0.1218 | Yes |
| 12 | Prkag1 | 1184 | 0.356 | 0.1350 | Yes |
| 13 | Pak4 | 1233 | 0.349 | 0.1462 | Yes |
| 14 | Rit1 | 1272 | 0.344 | 0.1581 | Yes |
| 15 | Smad2 | 1343 | 0.334 | 0.1668 | Yes |
| 16 | Prkcb | 1502 | 0.316 | 0.1670 | Yes |
| 17 | Tbk1 | 1566 | 0.306 | 0.1750 | Yes |
| 18 | Sla | 1733 | 0.287 | 0.1733 | No |
| 19 | Cdk1 | 1921 | 0.268 | 0.1689 | No |
| 20 | Cfl1 | 2107 | 0.250 | 0.1638 | No |
| 21 | Raf1 | 2450 | 0.214 | 0.1436 | No |
| 22 | Tsc2 | 2616 | 0.198 | 0.1380 | No |
| 23 | Hras | 2685 | 0.193 | 0.1406 | No |
| 24 | Itpr2 | 2745 | 0.188 | 0.1438 | No |
| 25 | Mapk9 | 2795 | 0.183 | 0.1477 | No |
| 26 | Il4 | 2861 | 0.178 | 0.1499 | No |
| 27 | Irak4 | 2960 | 0.169 | 0.1488 | No |
| 28 | Ube2d3 | 3106 | 0.156 | 0.1432 | No |
| 29 | Mapkap1 | 3111 | 0.156 | 0.1497 | No |
| 30 | Nfkbib | 3190 | 0.150 | 0.1495 | No |
| 31 | Prkar2a | 3338 | 0.138 | 0.1428 | No |
| 32 | Ripk1 | 3376 | 0.135 | 0.1456 | No |
| 33 | Nod1 | 3405 | 0.133 | 0.1490 | No |
| 34 | Pdk1 | 3622 | 0.117 | 0.1354 | No |
| 35 | Mapk8 | 3652 | 0.115 | 0.1380 | No |
| 36 | Tnfrsf1a | 3693 | 0.112 | 0.1394 | No |
| 37 | Mknk1 | 3728 | 0.109 | 0.1413 | No |
| 38 | Gsk3b | 4057 | 0.084 | 0.1165 | No |
| 39 | Arpc3 | 4351 | 0.060 | 0.0937 | No |
| 40 | Grb2 | 4392 | 0.057 | 0.0928 | No |
| 41 | Atf1 | 4486 | 0.051 | 0.0870 | No |
| 42 | Myd88 | 4636 | 0.039 | 0.0758 | No |
| 43 | Cab39l | 4850 | 0.022 | 0.0583 | No |
| 44 | Sqstm1 | 4937 | 0.016 | 0.0515 | No |
| 45 | Calr | 5156 | 0.000 | 0.0326 | No |
| 46 | Pikfyve | 5384 | -0.016 | 0.0136 | No |
| 47 | Csnk2b | 5405 | -0.018 | 0.0127 | No |
| 48 | Pten | 5803 | -0.048 | -0.0197 | No |
| 49 | Actr2 | 5835 | -0.050 | -0.0202 | No |
| 50 | Rac1 | 5931 | -0.057 | -0.0259 | No |
| 51 | Rps6ka3 | 5932 | -0.057 | -0.0234 | No |
| 52 | Pin1 | 5993 | -0.061 | -0.0259 | No |
| 53 | Ppp1ca | 6008 | -0.062 | -0.0243 | No |
| 54 | Grk2 | 6009 | -0.062 | -0.0216 | No |
| 55 | Stat2 | 6015 | -0.063 | -0.0192 | No |
| 56 | Pfn1 | 6016 | -0.063 | -0.0164 | No |
| 57 | Pla2g12a | 6684 | -0.117 | -0.0692 | No |
| 58 | Them4 | 7156 | -0.156 | -0.1032 | No |
| 59 | Ap2m1 | 7405 | -0.175 | -0.1170 | No |
| 60 | Arhgdia | 7724 | -0.202 | -0.1357 | No |
| 61 | Cdk2 | 7729 | -0.202 | -0.1271 | No |
| 62 | Cdkn1a | 7917 | -0.220 | -0.1336 | No |
| 63 | Map2k3 | 8056 | -0.230 | -0.1355 | No |
| 64 | Cdk4 | 8062 | -0.230 | -0.1257 | No |
| 65 | Ywhab | 8155 | -0.238 | -0.1232 | No |
| 66 | Dapp1 | 8253 | -0.245 | -0.1208 | No |
| 67 | Ecsit | 8303 | -0.249 | -0.1141 | No |
| 68 | Akt1s1 | 8316 | -0.250 | -0.1041 | No |
| 69 | Ddit3 | 8327 | -0.251 | -0.0939 | No |
| 70 | Acaca | 8841 | -0.300 | -0.1251 | No |
| 71 | Cab39 | 8850 | -0.300 | -0.1126 | No |
| 72 | Mknk2 | 8906 | -0.306 | -0.1039 | No |
| 73 | Rptor | 9064 | -0.323 | -0.1032 | No |
| 74 | Ppp2r1b | 9268 | -0.344 | -0.1057 | No |
| 75 | Cxcr4 | 9283 | -0.345 | -0.0917 | No |
| 76 | Rps6ka1 | 9359 | -0.351 | -0.0827 | No |
| 77 | Il2rg | 9388 | -0.353 | -0.0695 | No |
| 78 | Eif4e | 9656 | -0.381 | -0.0759 | No |
| 79 | Vav3 | 9824 | -0.402 | -0.0726 | No |
| 80 | Plcg1 | 9899 | -0.413 | -0.0608 | No |
| 81 | Traf2 | 9916 | -0.415 | -0.0439 | No |
| 82 | Nck1 | 9969 | -0.423 | -0.0297 | No |
| 83 | Ube2n | 9983 | -0.425 | -0.0121 | No |
| 84 | Mapk1 | 10622 | -0.522 | -0.0445 | No |
| 85 | Map3k7 | 10637 | -0.525 | -0.0225 | No |
| 86 | Slc2a1 | 10688 | -0.536 | -0.0032 | No |
| 87 | Gna14 | 10755 | -0.552 | 0.0154 | No |
| 88 | Tiam1 | 11032 | -0.631 | 0.0193 | No |
| 89 | Prkaa2 | 11196 | -0.695 | 0.0358 | No |

