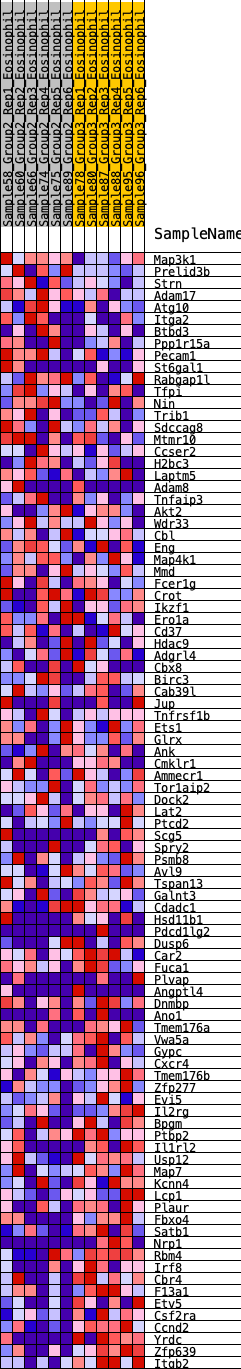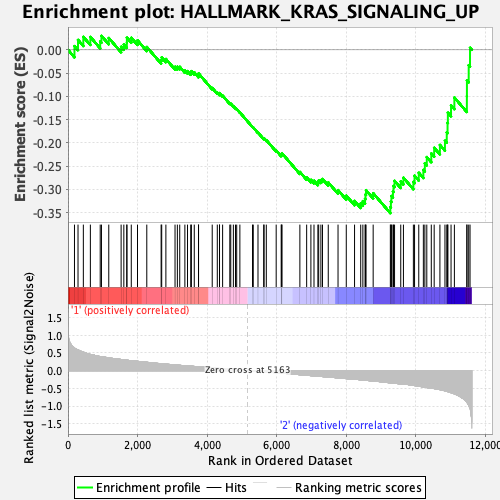
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.3510132 |
| Normalized Enrichment Score (NES) | -1.3961438 |
| Nominal p-value | 0.10526316 |
| FDR q-value | 0.33972698 |
| FWER p-Value | 0.619 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Map3k1 | 186 | 0.634 | 0.0080 | No |
| 2 | Prelid3b | 288 | 0.581 | 0.0213 | No |
| 3 | Strn | 441 | 0.519 | 0.0278 | No |
| 4 | Adam17 | 643 | 0.455 | 0.0277 | No |
| 5 | Atg10 | 926 | 0.394 | 0.0182 | No |
| 6 | Itga2 | 962 | 0.390 | 0.0300 | No |
| 7 | Btbd3 | 1172 | 0.357 | 0.0254 | No |
| 8 | Ppp1r15a | 1527 | 0.312 | 0.0066 | No |
| 9 | Pecam1 | 1605 | 0.302 | 0.0114 | No |
| 10 | St6gal1 | 1691 | 0.292 | 0.0151 | No |
| 11 | Rabgap1l | 1693 | 0.292 | 0.0261 | No |
| 12 | Tfpi | 1819 | 0.279 | 0.0259 | No |
| 13 | Nin | 1998 | 0.261 | 0.0203 | No |
| 14 | Trib1 | 2265 | 0.233 | 0.0061 | No |
| 15 | Sdccag8 | 2677 | 0.193 | -0.0223 | No |
| 16 | Mtmr10 | 2694 | 0.193 | -0.0163 | No |
| 17 | Ccser2 | 2814 | 0.182 | -0.0197 | No |
| 18 | H2bc3 | 3077 | 0.158 | -0.0364 | No |
| 19 | Laptm5 | 3144 | 0.154 | -0.0363 | No |
| 20 | Adam8 | 3212 | 0.147 | -0.0365 | No |
| 21 | Tnfaip3 | 3360 | 0.136 | -0.0442 | No |
| 22 | Akt2 | 3436 | 0.130 | -0.0457 | No |
| 23 | Wdr33 | 3529 | 0.124 | -0.0490 | No |
| 24 | Cbl | 3547 | 0.122 | -0.0458 | No |
| 25 | Eng | 3630 | 0.116 | -0.0485 | No |
| 26 | Map4k1 | 3752 | 0.108 | -0.0550 | No |
| 27 | Mmd | 3753 | 0.107 | -0.0509 | No |
| 28 | Fcer1g | 4146 | 0.077 | -0.0820 | No |
| 29 | Crot | 4292 | 0.066 | -0.0921 | No |
| 30 | Ikzf1 | 4355 | 0.060 | -0.0952 | No |
| 31 | Ero1a | 4356 | 0.059 | -0.0929 | No |
| 32 | Cd37 | 4445 | 0.053 | -0.0985 | No |
| 33 | Hdac9 | 4652 | 0.039 | -0.1150 | No |
| 34 | Adgrl4 | 4679 | 0.037 | -0.1158 | No |
| 35 | Cbx8 | 4754 | 0.031 | -0.1211 | No |
| 36 | Birc3 | 4815 | 0.026 | -0.1253 | No |
| 37 | Cab39l | 4850 | 0.022 | -0.1274 | No |
| 38 | Jup | 4941 | 0.015 | -0.1346 | No |
| 39 | Tnfrsf1b | 5304 | -0.011 | -0.1656 | No |
| 40 | Ets1 | 5326 | -0.012 | -0.1670 | No |
| 41 | Glrx | 5462 | -0.023 | -0.1779 | No |
| 42 | Ank | 5627 | -0.035 | -0.1908 | No |
| 43 | Cmklr1 | 5636 | -0.036 | -0.1901 | No |
| 44 | Ammecr1 | 5701 | -0.041 | -0.1941 | No |
| 45 | Tor1aip2 | 5985 | -0.061 | -0.2164 | No |
| 46 | Dock2 | 6129 | -0.072 | -0.2261 | No |
| 47 | Lat2 | 6142 | -0.072 | -0.2243 | No |
| 48 | Ptcd2 | 6151 | -0.073 | -0.2223 | No |
| 49 | Scg5 | 6666 | -0.115 | -0.2625 | No |
| 50 | Spry2 | 6861 | -0.132 | -0.2743 | No |
| 51 | Psmb8 | 6983 | -0.141 | -0.2794 | No |
| 52 | Avl9 | 7072 | -0.149 | -0.2814 | No |
| 53 | Tspan13 | 7184 | -0.158 | -0.2851 | No |
| 54 | Galnt3 | 7207 | -0.159 | -0.2809 | No |
| 55 | Cdadc1 | 7270 | -0.164 | -0.2801 | No |
| 56 | Hsd11b1 | 7316 | -0.168 | -0.2776 | No |
| 57 | Pdcd1lg2 | 7482 | -0.181 | -0.2850 | No |
| 58 | Dusp6 | 7763 | -0.205 | -0.3015 | No |
| 59 | Car2 | 8000 | -0.226 | -0.3134 | No |
| 60 | Fuca1 | 8238 | -0.244 | -0.3247 | No |
| 61 | Plvap | 8415 | -0.261 | -0.3301 | No |
| 62 | Angptl4 | 8477 | -0.268 | -0.3252 | No |
| 63 | Dnmbp | 8540 | -0.272 | -0.3203 | No |
| 64 | Ano1 | 8553 | -0.273 | -0.3109 | No |
| 65 | Tmem176a | 8568 | -0.275 | -0.3017 | No |
| 66 | Vwa5a | 8773 | -0.294 | -0.3082 | No |
| 67 | Gypc | 9267 | -0.344 | -0.3379 | Yes |
| 68 | Cxcr4 | 9283 | -0.345 | -0.3261 | Yes |
| 69 | Tmem176b | 9298 | -0.346 | -0.3142 | Yes |
| 70 | Zfp277 | 9342 | -0.350 | -0.3046 | Yes |
| 71 | Evi5 | 9358 | -0.351 | -0.2926 | Yes |
| 72 | Il2rg | 9388 | -0.353 | -0.2816 | Yes |
| 73 | Bpgm | 9566 | -0.374 | -0.2828 | Yes |
| 74 | Ptbp2 | 9643 | -0.381 | -0.2749 | Yes |
| 75 | Il1rl2 | 9929 | -0.417 | -0.2838 | Yes |
| 76 | Usp12 | 9963 | -0.422 | -0.2706 | Yes |
| 77 | Map7 | 10084 | -0.441 | -0.2642 | Yes |
| 78 | Kcnn4 | 10216 | -0.461 | -0.2581 | Yes |
| 79 | Lcp1 | 10259 | -0.470 | -0.2439 | Yes |
| 80 | Plaur | 10315 | -0.478 | -0.2305 | Yes |
| 81 | Fbxo4 | 10444 | -0.495 | -0.2227 | Yes |
| 82 | Satb1 | 10527 | -0.507 | -0.2106 | Yes |
| 83 | Nrp1 | 10692 | -0.537 | -0.2044 | Yes |
| 84 | Rbm4 | 10837 | -0.568 | -0.1953 | Yes |
| 85 | Irf8 | 10889 | -0.583 | -0.1776 | Yes |
| 86 | Cbr4 | 10912 | -0.590 | -0.1570 | Yes |
| 87 | F13a1 | 10921 | -0.594 | -0.1352 | Yes |
| 88 | Etv5 | 11012 | -0.624 | -0.1193 | Yes |
| 89 | Csf2ra | 11108 | -0.655 | -0.1026 | Yes |
| 90 | Ccnd2 | 11467 | -0.893 | -0.0997 | Yes |
| 91 | Yrdc | 11469 | -0.894 | -0.0658 | Yes |
| 92 | Zfp639 | 11520 | -0.974 | -0.0331 | Yes |
| 93 | Itgb2 | 11558 | -1.072 | 0.0044 | Yes |