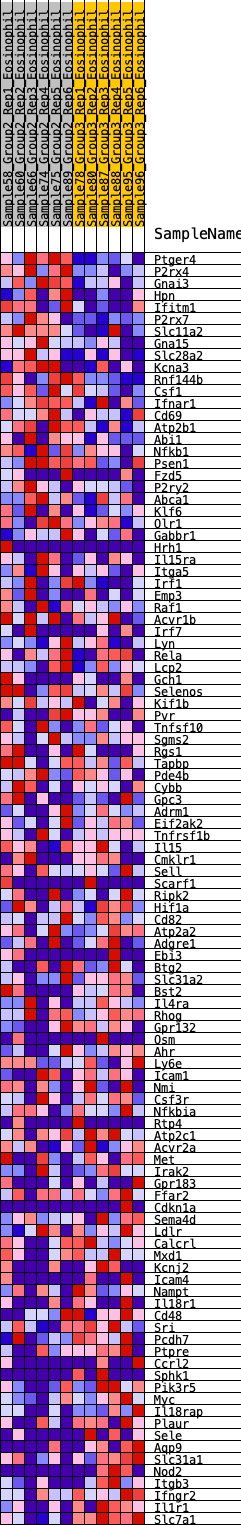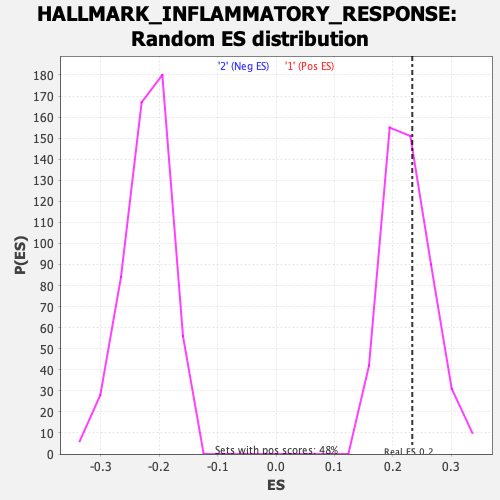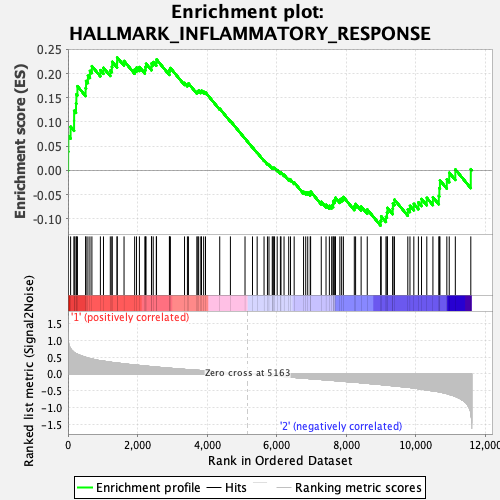
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.23329678 |
| Normalized Enrichment Score (NES) | 1.037875 |
| Nominal p-value | 0.3611691 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ptger4 | 4 | 1.207 | 0.0390 | Yes |
| 2 | P2rx4 | 10 | 0.998 | 0.0711 | Yes |
| 3 | Gnai3 | 75 | 0.760 | 0.0903 | Yes |
| 4 | Hpn | 175 | 0.639 | 0.1025 | Yes |
| 5 | Ifitm1 | 177 | 0.639 | 0.1232 | Yes |
| 6 | P2rx7 | 231 | 0.606 | 0.1384 | Yes |
| 7 | Slc11a2 | 241 | 0.603 | 0.1572 | Yes |
| 8 | Gna15 | 271 | 0.588 | 0.1739 | Yes |
| 9 | Slc28a2 | 506 | 0.496 | 0.1697 | Yes |
| 10 | Kcna3 | 519 | 0.494 | 0.1847 | Yes |
| 11 | Rnf144b | 573 | 0.475 | 0.1956 | Yes |
| 12 | Csf1 | 631 | 0.458 | 0.2056 | Yes |
| 13 | Ifnar1 | 687 | 0.445 | 0.2153 | Yes |
| 14 | Cd69 | 931 | 0.394 | 0.2070 | Yes |
| 15 | Atp2b1 | 1020 | 0.383 | 0.2118 | Yes |
| 16 | Abi1 | 1216 | 0.352 | 0.2064 | Yes |
| 17 | Nfkb1 | 1261 | 0.345 | 0.2138 | Yes |
| 18 | Psen1 | 1270 | 0.344 | 0.2243 | Yes |
| 19 | Fzd5 | 1411 | 0.326 | 0.2228 | Yes |
| 20 | P2ry2 | 1413 | 0.326 | 0.2333 | Yes |
| 21 | Abca1 | 1611 | 0.301 | 0.2260 | No |
| 22 | Klf6 | 1916 | 0.269 | 0.2083 | No |
| 23 | Olr1 | 1970 | 0.263 | 0.2123 | No |
| 24 | Gabbr1 | 2053 | 0.254 | 0.2134 | No |
| 25 | Hrh1 | 2210 | 0.238 | 0.2076 | No |
| 26 | Il15ra | 2228 | 0.237 | 0.2139 | No |
| 27 | Itga5 | 2243 | 0.235 | 0.2203 | No |
| 28 | Irf1 | 2400 | 0.219 | 0.2139 | No |
| 29 | Emp3 | 2402 | 0.218 | 0.2209 | No |
| 30 | Raf1 | 2450 | 0.214 | 0.2238 | No |
| 31 | Acvr1b | 2539 | 0.205 | 0.2229 | No |
| 32 | Irf7 | 2545 | 0.205 | 0.2291 | No |
| 33 | Lyn | 2915 | 0.173 | 0.2026 | No |
| 34 | Rela | 2918 | 0.173 | 0.2081 | No |
| 35 | Lcp2 | 2944 | 0.170 | 0.2115 | No |
| 36 | Gch1 | 3350 | 0.136 | 0.1807 | No |
| 37 | Selenos | 3433 | 0.131 | 0.1778 | No |
| 38 | Kif1b | 3462 | 0.129 | 0.1796 | No |
| 39 | Pvr | 3704 | 0.111 | 0.1623 | No |
| 40 | Tnfsf10 | 3732 | 0.109 | 0.1635 | No |
| 41 | Sgms2 | 3757 | 0.107 | 0.1649 | No |
| 42 | Rgs1 | 3819 | 0.102 | 0.1629 | No |
| 43 | Tapbp | 3838 | 0.101 | 0.1646 | No |
| 44 | Pde4b | 3900 | 0.097 | 0.1625 | No |
| 45 | Cybb | 3953 | 0.092 | 0.1610 | No |
| 46 | Gpc3 | 4362 | 0.059 | 0.1274 | No |
| 47 | Adrm1 | 4669 | 0.037 | 0.1020 | No |
| 48 | Eif2ak2 | 5089 | 0.005 | 0.0658 | No |
| 49 | Tnfrsf1b | 5304 | -0.011 | 0.0475 | No |
| 50 | Il15 | 5438 | -0.021 | 0.0367 | No |
| 51 | Cmklr1 | 5636 | -0.036 | 0.0207 | No |
| 52 | Sell | 5734 | -0.044 | 0.0137 | No |
| 53 | Scarf1 | 5778 | -0.047 | 0.0115 | No |
| 54 | Ripk2 | 5872 | -0.053 | 0.0051 | No |
| 55 | Hif1a | 5891 | -0.054 | 0.0053 | No |
| 56 | Cd82 | 5915 | -0.056 | 0.0051 | No |
| 57 | Atp2a2 | 5938 | -0.058 | 0.0051 | No |
| 58 | Adgre1 | 6014 | -0.063 | 0.0006 | No |
| 59 | Ebi3 | 6106 | -0.070 | -0.0050 | No |
| 60 | Btg2 | 6123 | -0.072 | -0.0040 | No |
| 61 | Slc31a2 | 6211 | -0.077 | -0.0091 | No |
| 62 | Bst2 | 6345 | -0.089 | -0.0177 | No |
| 63 | Il4ra | 6397 | -0.094 | -0.0191 | No |
| 64 | Rhog | 6502 | -0.102 | -0.0248 | No |
| 65 | Gpr132 | 6772 | -0.124 | -0.0442 | No |
| 66 | Osm | 6834 | -0.130 | -0.0452 | No |
| 67 | Ahr | 6892 | -0.134 | -0.0458 | No |
| 68 | Ly6e | 6963 | -0.140 | -0.0473 | No |
| 69 | Icam1 | 6975 | -0.141 | -0.0437 | No |
| 70 | Nmi | 7281 | -0.165 | -0.0648 | No |
| 71 | Csf3r | 7418 | -0.176 | -0.0709 | No |
| 72 | Nfkbia | 7514 | -0.184 | -0.0732 | No |
| 73 | Rtp4 | 7580 | -0.190 | -0.0726 | No |
| 74 | Atp2c1 | 7622 | -0.193 | -0.0699 | No |
| 75 | Acvr2a | 7625 | -0.193 | -0.0638 | No |
| 76 | Met | 7666 | -0.197 | -0.0609 | No |
| 77 | Irak2 | 7690 | -0.199 | -0.0564 | No |
| 78 | Gpr183 | 7816 | -0.210 | -0.0604 | No |
| 79 | Ffar2 | 7867 | -0.215 | -0.0577 | No |
| 80 | Cdkn1a | 7917 | -0.220 | -0.0548 | No |
| 81 | Sema4d | 8231 | -0.243 | -0.0741 | No |
| 82 | Ldlr | 8268 | -0.246 | -0.0692 | No |
| 83 | Calcrl | 8427 | -0.262 | -0.0744 | No |
| 84 | Mxd1 | 8603 | -0.278 | -0.0806 | No |
| 85 | Kcnj2 | 8988 | -0.314 | -0.1037 | No |
| 86 | Icam4 | 9004 | -0.316 | -0.0948 | No |
| 87 | Nampt | 9141 | -0.331 | -0.0958 | No |
| 88 | Il18r1 | 9170 | -0.336 | -0.0873 | No |
| 89 | Cd48 | 9183 | -0.337 | -0.0774 | No |
| 90 | Sri | 9335 | -0.350 | -0.0791 | No |
| 91 | Pcdh7 | 9341 | -0.350 | -0.0681 | No |
| 92 | Ptpre | 9385 | -0.353 | -0.0604 | No |
| 93 | Ccrl2 | 9769 | -0.392 | -0.0809 | No |
| 94 | Sphk1 | 9831 | -0.402 | -0.0731 | No |
| 95 | Pik3r5 | 9942 | -0.419 | -0.0690 | No |
| 96 | Myc | 10073 | -0.438 | -0.0660 | No |
| 97 | Il18rap | 10163 | -0.452 | -0.0590 | No |
| 98 | Plaur | 10315 | -0.478 | -0.0565 | No |
| 99 | Sele | 10491 | -0.502 | -0.0554 | No |
| 100 | Aqp9 | 10661 | -0.530 | -0.0528 | No |
| 101 | Slc31a1 | 10676 | -0.534 | -0.0366 | No |
| 102 | Nod2 | 10693 | -0.537 | -0.0205 | No |
| 103 | Itgb3 | 10891 | -0.583 | -0.0186 | No |
| 104 | Ifngr2 | 10960 | -0.608 | -0.0047 | No |
| 105 | Il1r1 | 11135 | -0.669 | 0.0020 | No |
| 106 | Slc7a1 | 11580 | -1.201 | 0.0025 | No |