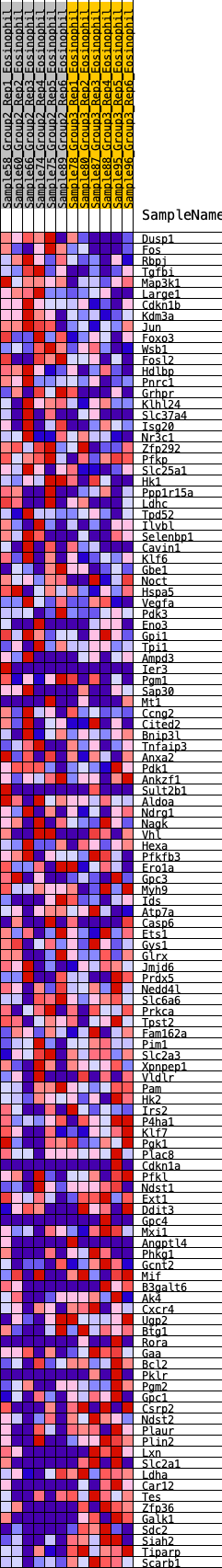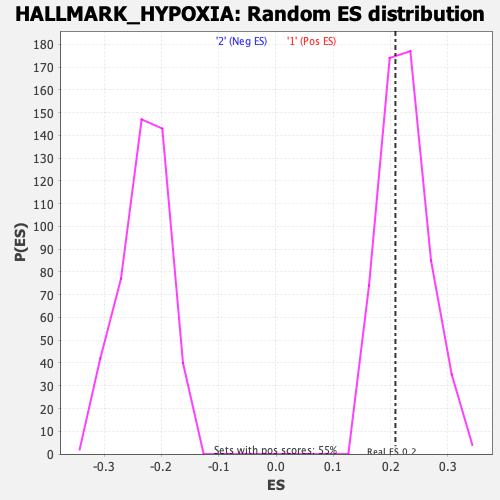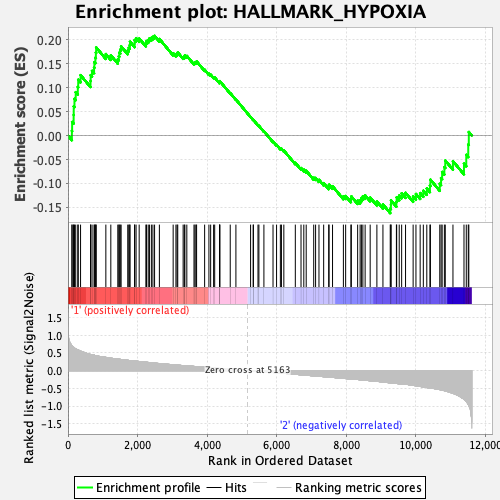
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.20833576 |
| Normalized Enrichment Score (NES) | 0.9271684 |
| Nominal p-value | 0.61566484 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dusp1 | 112 | 0.704 | 0.0098 | Yes |
| 2 | Fos | 119 | 0.695 | 0.0285 | Yes |
| 3 | Rbpj | 162 | 0.656 | 0.0431 | Yes |
| 4 | Tgfbi | 165 | 0.653 | 0.0610 | Yes |
| 5 | Map3k1 | 186 | 0.634 | 0.0768 | Yes |
| 6 | Large1 | 222 | 0.610 | 0.0907 | Yes |
| 7 | Cdkn1b | 282 | 0.583 | 0.1017 | Yes |
| 8 | Kdm3a | 294 | 0.578 | 0.1168 | Yes |
| 9 | Jun | 361 | 0.548 | 0.1262 | Yes |
| 10 | Foxo3 | 647 | 0.455 | 0.1140 | Yes |
| 11 | Wsb1 | 652 | 0.453 | 0.1262 | Yes |
| 12 | Fosl2 | 691 | 0.444 | 0.1352 | Yes |
| 13 | Hdlbp | 750 | 0.432 | 0.1422 | Yes |
| 14 | Pnrc1 | 759 | 0.431 | 0.1534 | Yes |
| 15 | Grhpr | 787 | 0.422 | 0.1628 | Yes |
| 16 | Klhl24 | 801 | 0.420 | 0.1733 | Yes |
| 17 | Slc37a4 | 810 | 0.418 | 0.1842 | Yes |
| 18 | Isg20 | 1088 | 0.371 | 0.1703 | Yes |
| 19 | Nr3c1 | 1231 | 0.350 | 0.1677 | Yes |
| 20 | Zfp292 | 1432 | 0.324 | 0.1593 | Yes |
| 21 | Pfkp | 1461 | 0.320 | 0.1657 | Yes |
| 22 | Slc25a1 | 1475 | 0.318 | 0.1734 | Yes |
| 23 | Hk1 | 1503 | 0.315 | 0.1798 | Yes |
| 24 | Ppp1r15a | 1527 | 0.312 | 0.1864 | Yes |
| 25 | Ldhc | 1718 | 0.289 | 0.1779 | Yes |
| 26 | Tpd52 | 1747 | 0.285 | 0.1834 | Yes |
| 27 | Ilvbl | 1775 | 0.283 | 0.1889 | Yes |
| 28 | Selenbp1 | 1782 | 0.282 | 0.1962 | Yes |
| 29 | Cavin1 | 1915 | 0.269 | 0.1921 | Yes |
| 30 | Klf6 | 1916 | 0.269 | 0.1996 | Yes |
| 31 | Gbe1 | 1957 | 0.264 | 0.2034 | Yes |
| 32 | Noct | 2046 | 0.256 | 0.2029 | Yes |
| 33 | Hspa5 | 2239 | 0.236 | 0.1927 | Yes |
| 34 | Vegfa | 2258 | 0.234 | 0.1976 | Yes |
| 35 | Pdk3 | 2306 | 0.229 | 0.1998 | Yes |
| 36 | Eno3 | 2341 | 0.224 | 0.2031 | Yes |
| 37 | Gpi1 | 2397 | 0.219 | 0.2044 | Yes |
| 38 | Tpi1 | 2441 | 0.215 | 0.2066 | Yes |
| 39 | Ampd3 | 2489 | 0.211 | 0.2083 | Yes |
| 40 | Ier3 | 2627 | 0.197 | 0.2019 | No |
| 41 | Pgm1 | 3023 | 0.163 | 0.1720 | No |
| 42 | Sap30 | 3103 | 0.157 | 0.1695 | No |
| 43 | Mt1 | 3136 | 0.154 | 0.1710 | No |
| 44 | Ccng2 | 3155 | 0.153 | 0.1737 | No |
| 45 | Cited2 | 3310 | 0.140 | 0.1641 | No |
| 46 | Bnip3l | 3345 | 0.137 | 0.1650 | No |
| 47 | Tnfaip3 | 3360 | 0.136 | 0.1675 | No |
| 48 | Anxa2 | 3416 | 0.132 | 0.1664 | No |
| 49 | Pdk1 | 3622 | 0.117 | 0.1518 | No |
| 50 | Ankzf1 | 3650 | 0.115 | 0.1526 | No |
| 51 | Sult2b1 | 3680 | 0.113 | 0.1532 | No |
| 52 | Aldoa | 3697 | 0.112 | 0.1549 | No |
| 53 | Ndrg1 | 3928 | 0.094 | 0.1375 | No |
| 54 | Nagk | 4051 | 0.085 | 0.1292 | No |
| 55 | Vhl | 4101 | 0.081 | 0.1272 | No |
| 56 | Hexa | 4186 | 0.074 | 0.1219 | No |
| 57 | Pfkfb3 | 4217 | 0.072 | 0.1213 | No |
| 58 | Ero1a | 4356 | 0.059 | 0.1110 | No |
| 59 | Gpc3 | 4362 | 0.059 | 0.1122 | No |
| 60 | Myh9 | 4364 | 0.059 | 0.1137 | No |
| 61 | Ids | 4664 | 0.038 | 0.0887 | No |
| 62 | Atp7a | 4827 | 0.025 | 0.0753 | No |
| 63 | Casp6 | 5249 | -0.006 | 0.0388 | No |
| 64 | Ets1 | 5326 | -0.012 | 0.0326 | No |
| 65 | Gys1 | 5328 | -0.012 | 0.0328 | No |
| 66 | Glrx | 5462 | -0.023 | 0.0219 | No |
| 67 | Jmjd6 | 5488 | -0.024 | 0.0204 | No |
| 68 | Prdx5 | 5632 | -0.036 | 0.0089 | No |
| 69 | Nedd4l | 5894 | -0.054 | -0.0123 | No |
| 70 | Slc6a6 | 5996 | -0.062 | -0.0194 | No |
| 71 | Prkca | 6105 | -0.070 | -0.0268 | No |
| 72 | Tpst2 | 6136 | -0.072 | -0.0275 | No |
| 73 | Fam162a | 6205 | -0.077 | -0.0312 | No |
| 74 | Pim1 | 6535 | -0.105 | -0.0570 | No |
| 75 | Slc2a3 | 6699 | -0.119 | -0.0678 | No |
| 76 | Xpnpep1 | 6779 | -0.125 | -0.0713 | No |
| 77 | Vldlr | 6848 | -0.131 | -0.0735 | No |
| 78 | Pam | 7064 | -0.148 | -0.0881 | No |
| 79 | Hk2 | 7118 | -0.153 | -0.0885 | No |
| 80 | Irs2 | 7215 | -0.159 | -0.0925 | No |
| 81 | P4ha1 | 7350 | -0.171 | -0.0994 | No |
| 82 | Klf7 | 7496 | -0.181 | -0.1070 | No |
| 83 | Pgk1 | 7504 | -0.183 | -0.1025 | No |
| 84 | Plac8 | 7605 | -0.192 | -0.1059 | No |
| 85 | Cdkn1a | 7917 | -0.220 | -0.1269 | No |
| 86 | Pfkl | 7982 | -0.224 | -0.1263 | No |
| 87 | Ndst1 | 8133 | -0.236 | -0.1328 | No |
| 88 | Ext1 | 8145 | -0.237 | -0.1271 | No |
| 89 | Ddit3 | 8327 | -0.251 | -0.1359 | No |
| 90 | Gpc4 | 8398 | -0.259 | -0.1349 | No |
| 91 | Mxi1 | 8435 | -0.263 | -0.1307 | No |
| 92 | Angptl4 | 8477 | -0.268 | -0.1268 | No |
| 93 | Phkg1 | 8542 | -0.272 | -0.1249 | No |
| 94 | Gcnt2 | 8687 | -0.287 | -0.1295 | No |
| 95 | Mif | 8880 | -0.303 | -0.1378 | No |
| 96 | B3galt6 | 9054 | -0.322 | -0.1439 | No |
| 97 | Ak4 | 9264 | -0.344 | -0.1525 | No |
| 98 | Cxcr4 | 9283 | -0.345 | -0.1446 | No |
| 99 | Ugp2 | 9286 | -0.345 | -0.1352 | No |
| 100 | Btg1 | 9441 | -0.362 | -0.1385 | No |
| 101 | Rora | 9449 | -0.362 | -0.1291 | No |
| 102 | Gaa | 9522 | -0.371 | -0.1251 | No |
| 103 | Bcl2 | 9592 | -0.377 | -0.1206 | No |
| 104 | Pklr | 9704 | -0.386 | -0.1196 | No |
| 105 | Pgm2 | 9922 | -0.416 | -0.1270 | No |
| 106 | Gpc1 | 10003 | -0.428 | -0.1221 | No |
| 107 | Csrp2 | 10124 | -0.446 | -0.1201 | No |
| 108 | Ndst2 | 10215 | -0.461 | -0.1152 | No |
| 109 | Plaur | 10315 | -0.478 | -0.1106 | No |
| 110 | Plin2 | 10405 | -0.492 | -0.1047 | No |
| 111 | Lxn | 10419 | -0.493 | -0.0921 | No |
| 112 | Slc2a1 | 10688 | -0.536 | -0.1006 | No |
| 113 | Ldha | 10725 | -0.544 | -0.0887 | No |
| 114 | Car12 | 10756 | -0.552 | -0.0760 | No |
| 115 | Tes | 10820 | -0.566 | -0.0658 | No |
| 116 | Zfp36 | 10846 | -0.570 | -0.0521 | No |
| 117 | Galk1 | 11067 | -0.641 | -0.0535 | No |
| 118 | Sdc2 | 11384 | -0.811 | -0.0586 | No |
| 119 | Siah2 | 11450 | -0.865 | -0.0403 | No |
| 120 | Tiparp | 11507 | -0.956 | -0.0186 | No |
| 121 | Scarb1 | 11523 | -0.989 | 0.0075 | No |