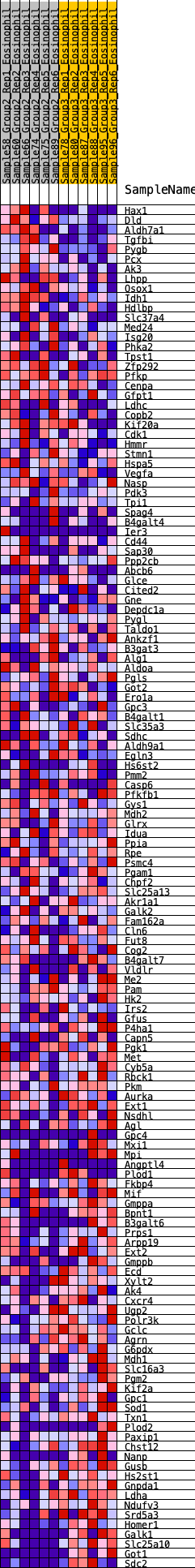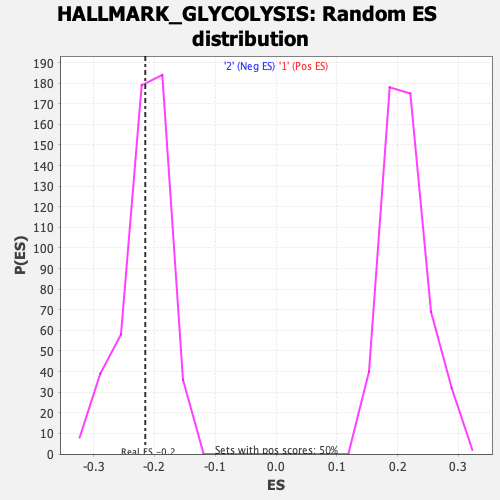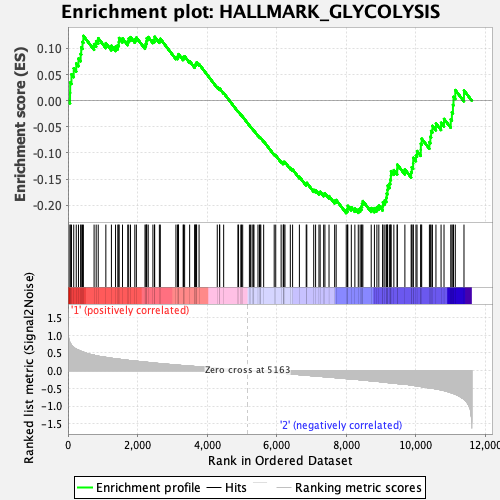
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_GLYCOLYSIS |
| Enrichment Score (ES) | -0.21506467 |
| Normalized Enrichment Score (NES) | -1.0012841 |
| Nominal p-value | 0.43452382 |
| FDR q-value | 0.7701581 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hax1 | 59 | 0.787 | 0.0152 | No |
| 2 | Dld | 63 | 0.777 | 0.0351 | No |
| 3 | Aldh7a1 | 100 | 0.719 | 0.0505 | No |
| 4 | Tgfbi | 165 | 0.653 | 0.0618 | No |
| 5 | Pygb | 237 | 0.604 | 0.0713 | No |
| 6 | Pcx | 301 | 0.575 | 0.0807 | No |
| 7 | Ak3 | 364 | 0.547 | 0.0894 | No |
| 8 | Lhpp | 380 | 0.541 | 0.1021 | No |
| 9 | Qsox1 | 417 | 0.528 | 0.1126 | No |
| 10 | Idh1 | 442 | 0.519 | 0.1240 | No |
| 11 | Hdlbp | 750 | 0.432 | 0.1084 | No |
| 12 | Slc37a4 | 810 | 0.418 | 0.1140 | No |
| 13 | Med24 | 869 | 0.405 | 0.1195 | No |
| 14 | Isg20 | 1088 | 0.371 | 0.1101 | No |
| 15 | Phka2 | 1248 | 0.347 | 0.1052 | No |
| 16 | Tpst1 | 1367 | 0.331 | 0.1034 | No |
| 17 | Zfp292 | 1432 | 0.324 | 0.1062 | No |
| 18 | Pfkp | 1461 | 0.320 | 0.1121 | No |
| 19 | Cenpa | 1467 | 0.319 | 0.1199 | No |
| 20 | Gfpt1 | 1567 | 0.306 | 0.1192 | No |
| 21 | Ldhc | 1718 | 0.289 | 0.1136 | No |
| 22 | Copb2 | 1745 | 0.286 | 0.1187 | No |
| 23 | Kif20a | 1797 | 0.281 | 0.1215 | No |
| 24 | Cdk1 | 1921 | 0.268 | 0.1177 | No |
| 25 | Hmmr | 1965 | 0.264 | 0.1208 | No |
| 26 | Stmn1 | 2214 | 0.238 | 0.1053 | No |
| 27 | Hspa5 | 2239 | 0.236 | 0.1093 | No |
| 28 | Vegfa | 2258 | 0.234 | 0.1138 | No |
| 29 | Nasp | 2264 | 0.233 | 0.1194 | No |
| 30 | Pdk3 | 2306 | 0.229 | 0.1218 | No |
| 31 | Tpi1 | 2441 | 0.215 | 0.1156 | No |
| 32 | Spag4 | 2486 | 0.211 | 0.1172 | No |
| 33 | B4galt4 | 2492 | 0.210 | 0.1223 | No |
| 34 | Ier3 | 2627 | 0.197 | 0.1157 | No |
| 35 | Cd44 | 2654 | 0.195 | 0.1184 | No |
| 36 | Sap30 | 3103 | 0.157 | 0.0834 | No |
| 37 | Ppp2cb | 3150 | 0.153 | 0.0834 | No |
| 38 | Abcb6 | 3168 | 0.152 | 0.0858 | No |
| 39 | Glce | 3177 | 0.151 | 0.0890 | No |
| 40 | Cited2 | 3310 | 0.140 | 0.0812 | No |
| 41 | Gne | 3321 | 0.140 | 0.0839 | No |
| 42 | Depdc1a | 3353 | 0.136 | 0.0847 | No |
| 43 | Pygl | 3497 | 0.126 | 0.0755 | No |
| 44 | Taldo1 | 3643 | 0.115 | 0.0659 | No |
| 45 | Ankzf1 | 3650 | 0.115 | 0.0683 | No |
| 46 | B3gat3 | 3670 | 0.114 | 0.0696 | No |
| 47 | Alg1 | 3686 | 0.112 | 0.0712 | No |
| 48 | Aldoa | 3697 | 0.112 | 0.0732 | No |
| 49 | Pgls | 3768 | 0.107 | 0.0699 | No |
| 50 | Got2 | 4291 | 0.066 | 0.0261 | No |
| 51 | Ero1a | 4356 | 0.059 | 0.0220 | No |
| 52 | Gpc3 | 4362 | 0.059 | 0.0231 | No |
| 53 | B4galt1 | 4475 | 0.052 | 0.0147 | No |
| 54 | Slc35a3 | 4885 | 0.019 | -0.0205 | No |
| 55 | Sdhc | 4903 | 0.018 | -0.0215 | No |
| 56 | Aldh9a1 | 4969 | 0.013 | -0.0268 | No |
| 57 | Egln3 | 4989 | 0.012 | -0.0282 | No |
| 58 | Hs6st2 | 5022 | 0.009 | -0.0307 | No |
| 59 | Pmm2 | 5215 | -0.003 | -0.0474 | No |
| 60 | Casp6 | 5249 | -0.006 | -0.0501 | No |
| 61 | Pfkfb1 | 5293 | -0.010 | -0.0536 | No |
| 62 | Gys1 | 5328 | -0.012 | -0.0562 | No |
| 63 | Mdh2 | 5346 | -0.013 | -0.0574 | No |
| 64 | Glrx | 5462 | -0.023 | -0.0668 | No |
| 65 | Idua | 5505 | -0.026 | -0.0698 | No |
| 66 | Ppia | 5524 | -0.027 | -0.0707 | No |
| 67 | Rpe | 5543 | -0.029 | -0.0715 | No |
| 68 | Psmc4 | 5625 | -0.035 | -0.0777 | No |
| 69 | Pgam1 | 5928 | -0.057 | -0.1025 | No |
| 70 | Chpf2 | 5972 | -0.060 | -0.1047 | No |
| 71 | Slc25a13 | 6127 | -0.072 | -0.1163 | No |
| 72 | Akr1a1 | 6187 | -0.076 | -0.1195 | No |
| 73 | Galk2 | 6200 | -0.077 | -0.1185 | No |
| 74 | Fam162a | 6205 | -0.077 | -0.1169 | No |
| 75 | Cln6 | 6237 | -0.079 | -0.1175 | No |
| 76 | Fut8 | 6391 | -0.093 | -0.1285 | No |
| 77 | Cog2 | 6457 | -0.099 | -0.1316 | No |
| 78 | B4galt7 | 6652 | -0.114 | -0.1455 | No |
| 79 | Vldlr | 6848 | -0.131 | -0.1591 | No |
| 80 | Me2 | 6865 | -0.132 | -0.1571 | No |
| 81 | Pam | 7064 | -0.148 | -0.1705 | No |
| 82 | Hk2 | 7118 | -0.153 | -0.1712 | No |
| 83 | Irs2 | 7215 | -0.159 | -0.1754 | No |
| 84 | Gfus | 7249 | -0.162 | -0.1741 | No |
| 85 | P4ha1 | 7350 | -0.171 | -0.1784 | No |
| 86 | Capn5 | 7386 | -0.173 | -0.1770 | No |
| 87 | Pgk1 | 7504 | -0.183 | -0.1825 | No |
| 88 | Met | 7666 | -0.197 | -0.1914 | No |
| 89 | Cyb5a | 7709 | -0.201 | -0.1899 | No |
| 90 | Rbck1 | 7999 | -0.226 | -0.2092 | Yes |
| 91 | Pkm | 8037 | -0.229 | -0.2065 | Yes |
| 92 | Aurka | 8043 | -0.229 | -0.2010 | Yes |
| 93 | Ext1 | 8145 | -0.237 | -0.2037 | Yes |
| 94 | Nsdhl | 8247 | -0.244 | -0.2062 | Yes |
| 95 | Agl | 8345 | -0.253 | -0.2081 | Yes |
| 96 | Gpc4 | 8398 | -0.259 | -0.2059 | Yes |
| 97 | Mxi1 | 8435 | -0.263 | -0.2023 | Yes |
| 98 | Mpi | 8453 | -0.265 | -0.1969 | Yes |
| 99 | Angptl4 | 8477 | -0.268 | -0.1920 | Yes |
| 100 | Plod1 | 8717 | -0.289 | -0.2053 | Yes |
| 101 | Fkbp4 | 8809 | -0.298 | -0.2056 | Yes |
| 102 | Mif | 8880 | -0.303 | -0.2038 | Yes |
| 103 | Gmppa | 8936 | -0.309 | -0.2006 | Yes |
| 104 | Bpnt1 | 9046 | -0.321 | -0.2018 | Yes |
| 105 | B3galt6 | 9054 | -0.322 | -0.1941 | Yes |
| 106 | Prps1 | 9109 | -0.326 | -0.1903 | Yes |
| 107 | Arpp19 | 9151 | -0.333 | -0.1853 | Yes |
| 108 | Ext2 | 9162 | -0.335 | -0.1775 | Yes |
| 109 | Gmppb | 9185 | -0.337 | -0.1707 | Yes |
| 110 | Ecd | 9191 | -0.337 | -0.1624 | Yes |
| 111 | Xylt2 | 9247 | -0.342 | -0.1584 | Yes |
| 112 | Ak4 | 9264 | -0.344 | -0.1509 | Yes |
| 113 | Cxcr4 | 9283 | -0.345 | -0.1435 | Yes |
| 114 | Ugp2 | 9286 | -0.345 | -0.1348 | Yes |
| 115 | Polr3k | 9368 | -0.352 | -0.1327 | Yes |
| 116 | Gclc | 9464 | -0.365 | -0.1316 | Yes |
| 117 | Agrn | 9466 | -0.365 | -0.1222 | Yes |
| 118 | G6pdx | 9685 | -0.384 | -0.1313 | Yes |
| 119 | Mdh1 | 9865 | -0.407 | -0.1364 | Yes |
| 120 | Slc16a3 | 9883 | -0.410 | -0.1272 | Yes |
| 121 | Pgm2 | 9922 | -0.416 | -0.1198 | Yes |
| 122 | Kif2a | 9925 | -0.416 | -0.1092 | Yes |
| 123 | Gpc1 | 10003 | -0.428 | -0.1048 | Yes |
| 124 | Sod1 | 10034 | -0.432 | -0.0962 | Yes |
| 125 | Txn1 | 10142 | -0.450 | -0.0939 | Yes |
| 126 | Plod2 | 10144 | -0.450 | -0.0824 | Yes |
| 127 | Paxip1 | 10168 | -0.453 | -0.0727 | Yes |
| 128 | Chst12 | 10390 | -0.489 | -0.0793 | Yes |
| 129 | Nanp | 10417 | -0.493 | -0.0688 | Yes |
| 130 | Gusb | 10439 | -0.495 | -0.0578 | Yes |
| 131 | Hs2st1 | 10477 | -0.500 | -0.0481 | Yes |
| 132 | Gnpda1 | 10578 | -0.516 | -0.0435 | Yes |
| 133 | Ldha | 10725 | -0.544 | -0.0421 | Yes |
| 134 | Ndufv3 | 10810 | -0.564 | -0.0349 | Yes |
| 135 | Srd5a3 | 11004 | -0.621 | -0.0356 | Yes |
| 136 | Homer1 | 11037 | -0.632 | -0.0221 | Yes |
| 137 | Galk1 | 11067 | -0.641 | -0.0080 | Yes |
| 138 | Slc25a10 | 11083 | -0.646 | 0.0074 | Yes |
| 139 | Got1 | 11134 | -0.669 | 0.0203 | Yes |
| 140 | Sdc2 | 11384 | -0.811 | 0.0196 | Yes |