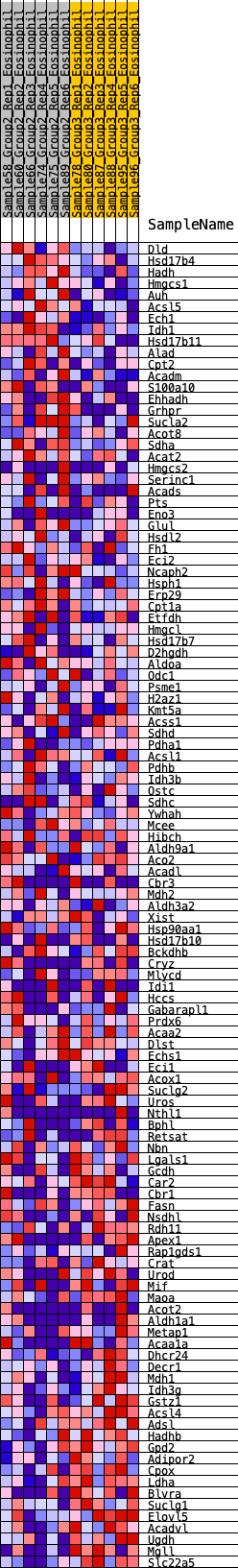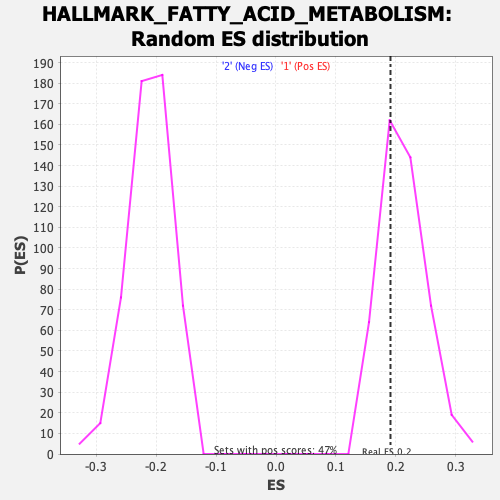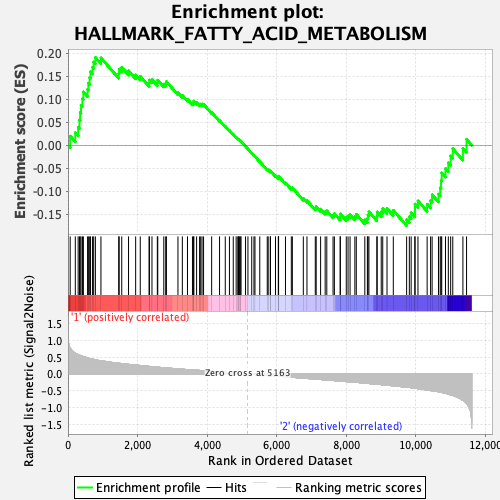
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | 0.1908424 |
| Normalized Enrichment Score (NES) | 0.9007181 |
| Nominal p-value | 0.6980728 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dld | 63 | 0.777 | 0.0201 | Yes |
| 2 | Hsd17b4 | 210 | 0.617 | 0.0277 | Yes |
| 3 | Hadh | 292 | 0.579 | 0.0397 | Yes |
| 4 | Hmgcs1 | 330 | 0.564 | 0.0551 | Yes |
| 5 | Auh | 350 | 0.553 | 0.0716 | Yes |
| 6 | Acsl5 | 375 | 0.543 | 0.0874 | Yes |
| 7 | Ech1 | 416 | 0.529 | 0.1013 | Yes |
| 8 | Idh1 | 442 | 0.519 | 0.1162 | Yes |
| 9 | Hsd17b11 | 563 | 0.479 | 0.1215 | Yes |
| 10 | Alad | 586 | 0.470 | 0.1351 | Yes |
| 11 | Cpt2 | 623 | 0.459 | 0.1471 | Yes |
| 12 | Acadm | 648 | 0.454 | 0.1599 | Yes |
| 13 | S100a10 | 706 | 0.441 | 0.1695 | Yes |
| 14 | Ehhadh | 738 | 0.436 | 0.1811 | Yes |
| 15 | Grhpr | 787 | 0.422 | 0.1908 | Yes |
| 16 | Sucla2 | 947 | 0.392 | 0.1899 | No |
| 17 | Acot8 | 1459 | 0.320 | 0.1560 | No |
| 18 | Sdha | 1470 | 0.319 | 0.1656 | No |
| 19 | Acat2 | 1545 | 0.310 | 0.1694 | No |
| 20 | Hmgcs2 | 1740 | 0.286 | 0.1619 | No |
| 21 | Serinc1 | 1945 | 0.266 | 0.1529 | No |
| 22 | Acads | 2077 | 0.253 | 0.1499 | No |
| 23 | Pts | 2331 | 0.225 | 0.1353 | No |
| 24 | Eno3 | 2341 | 0.224 | 0.1419 | No |
| 25 | Glul | 2414 | 0.217 | 0.1427 | No |
| 26 | Hsdl2 | 2568 | 0.202 | 0.1361 | No |
| 27 | Fh1 | 2581 | 0.201 | 0.1417 | No |
| 28 | Eci2 | 2754 | 0.188 | 0.1329 | No |
| 29 | Ncaph2 | 2809 | 0.183 | 0.1342 | No |
| 30 | Hsph1 | 2829 | 0.181 | 0.1385 | No |
| 31 | Erp29 | 3161 | 0.152 | 0.1147 | No |
| 32 | Cpt1a | 3287 | 0.142 | 0.1085 | No |
| 33 | Etfdh | 3435 | 0.130 | 0.1000 | No |
| 34 | Hmgcl | 3578 | 0.120 | 0.0916 | No |
| 35 | Hsd17b7 | 3612 | 0.117 | 0.0926 | No |
| 36 | D2hgdh | 3616 | 0.117 | 0.0962 | No |
| 37 | Aldoa | 3697 | 0.112 | 0.0929 | No |
| 38 | Odc1 | 3783 | 0.105 | 0.0890 | No |
| 39 | Psme1 | 3815 | 0.103 | 0.0897 | No |
| 40 | H2az1 | 3853 | 0.100 | 0.0897 | No |
| 41 | Kmt5a | 3897 | 0.097 | 0.0892 | No |
| 42 | Acss1 | 4130 | 0.078 | 0.0715 | No |
| 43 | Sdhd | 4357 | 0.059 | 0.0538 | No |
| 44 | Pdha1 | 4521 | 0.048 | 0.0413 | No |
| 45 | Acsl1 | 4641 | 0.039 | 0.0322 | No |
| 46 | Pdhb | 4751 | 0.032 | 0.0237 | No |
| 47 | Idh3b | 4834 | 0.024 | 0.0174 | No |
| 48 | Ostc | 4881 | 0.019 | 0.0140 | No |
| 49 | Sdhc | 4903 | 0.018 | 0.0128 | No |
| 50 | Ywhah | 4909 | 0.018 | 0.0130 | No |
| 51 | Mcee | 4933 | 0.016 | 0.0115 | No |
| 52 | Hibch | 4968 | 0.013 | 0.0089 | No |
| 53 | Aldh9a1 | 4969 | 0.013 | 0.0094 | No |
| 54 | Aco2 | 5104 | 0.003 | -0.0022 | No |
| 55 | Acadl | 5173 | -0.000 | -0.0081 | No |
| 56 | Cbr3 | 5282 | -0.009 | -0.0172 | No |
| 57 | Mdh2 | 5346 | -0.013 | -0.0222 | No |
| 58 | Aldh3a2 | 5375 | -0.015 | -0.0241 | No |
| 59 | Xist | 5513 | -0.026 | -0.0352 | No |
| 60 | Hsp90aa1 | 5728 | -0.044 | -0.0524 | No |
| 61 | Hsd17b10 | 5755 | -0.045 | -0.0532 | No |
| 62 | Bckdhb | 5817 | -0.049 | -0.0569 | No |
| 63 | Cryz | 5819 | -0.049 | -0.0553 | No |
| 64 | Mlycd | 5965 | -0.060 | -0.0660 | No |
| 65 | Idi1 | 6047 | -0.066 | -0.0709 | No |
| 66 | Hccs | 6050 | -0.066 | -0.0689 | No |
| 67 | Gabarapl1 | 6054 | -0.066 | -0.0670 | No |
| 68 | Prdx6 | 6252 | -0.081 | -0.0814 | No |
| 69 | Acaa2 | 6420 | -0.096 | -0.0928 | No |
| 70 | Dlst | 6450 | -0.099 | -0.0921 | No |
| 71 | Echs1 | 6765 | -0.124 | -0.1153 | No |
| 72 | Eci1 | 6870 | -0.133 | -0.1200 | No |
| 73 | Acox1 | 7109 | -0.152 | -0.1357 | No |
| 74 | Suclg2 | 7135 | -0.154 | -0.1328 | No |
| 75 | Uros | 7262 | -0.164 | -0.1384 | No |
| 76 | Nthl1 | 7393 | -0.174 | -0.1439 | No |
| 77 | Bphl | 7440 | -0.178 | -0.1421 | No |
| 78 | Retsat | 7620 | -0.193 | -0.1513 | No |
| 79 | Nbn | 7658 | -0.196 | -0.1481 | No |
| 80 | Lgals1 | 7825 | -0.211 | -0.1556 | No |
| 81 | Gcdh | 7829 | -0.211 | -0.1489 | No |
| 82 | Car2 | 8000 | -0.226 | -0.1562 | No |
| 83 | Cbr1 | 8051 | -0.230 | -0.1530 | No |
| 84 | Fasn | 8109 | -0.234 | -0.1503 | No |
| 85 | Nsdhl | 8247 | -0.244 | -0.1541 | No |
| 86 | Rdh11 | 8288 | -0.248 | -0.1495 | No |
| 87 | Apex1 | 8533 | -0.272 | -0.1618 | No |
| 88 | Rap1gds1 | 8607 | -0.278 | -0.1590 | No |
| 89 | Crat | 8623 | -0.279 | -0.1511 | No |
| 90 | Urod | 8651 | -0.283 | -0.1441 | No |
| 91 | Mif | 8880 | -0.303 | -0.1540 | No |
| 92 | Maoa | 8887 | -0.305 | -0.1445 | No |
| 93 | Acot2 | 9005 | -0.316 | -0.1443 | No |
| 94 | Aldh1a1 | 9048 | -0.322 | -0.1373 | No |
| 95 | Metap1 | 9171 | -0.336 | -0.1369 | No |
| 96 | Acaa1a | 9352 | -0.350 | -0.1410 | No |
| 97 | Dhcr24 | 9735 | -0.389 | -0.1615 | No |
| 98 | Decr1 | 9813 | -0.400 | -0.1550 | No |
| 99 | Mdh1 | 9865 | -0.407 | -0.1460 | No |
| 100 | Idh3g | 9974 | -0.424 | -0.1415 | No |
| 101 | Gstz1 | 9975 | -0.424 | -0.1275 | No |
| 102 | Acsl4 | 10061 | -0.437 | -0.1205 | No |
| 103 | Adsl | 10325 | -0.479 | -0.1276 | No |
| 104 | Hadhb | 10424 | -0.494 | -0.1199 | No |
| 105 | Gpd2 | 10468 | -0.499 | -0.1072 | No |
| 106 | Adipor2 | 10651 | -0.528 | -0.1057 | No |
| 107 | Cpox | 10700 | -0.539 | -0.0921 | No |
| 108 | Ldha | 10725 | -0.544 | -0.0763 | No |
| 109 | Blvra | 10744 | -0.549 | -0.0598 | No |
| 110 | Suclg1 | 10852 | -0.571 | -0.0503 | No |
| 111 | Elovl5 | 10934 | -0.600 | -0.0376 | No |
| 112 | Acadvl | 11001 | -0.620 | -0.0230 | No |
| 113 | Ugdh | 11062 | -0.640 | -0.0072 | No |
| 114 | Mgll | 11353 | -0.785 | -0.0066 | No |
| 115 | Slc22a5 | 11458 | -0.873 | 0.0131 | No |