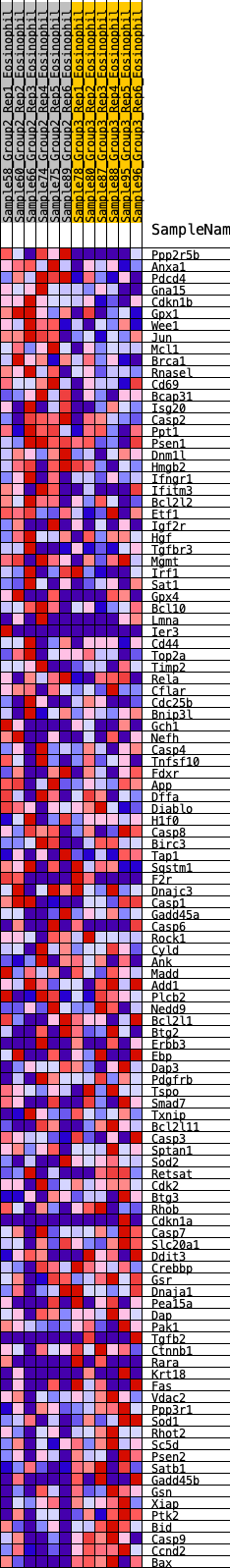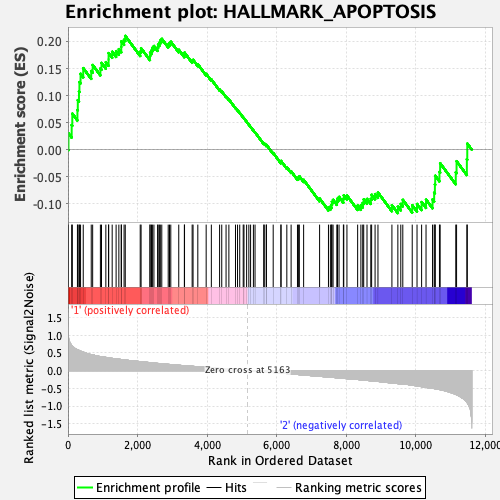
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.21028164 |
| Normalized Enrichment Score (NES) | 0.98303896 |
| Nominal p-value | 0.4612326 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ppp2r5b | 14 | 0.956 | 0.0302 | Yes |
| 2 | Anxa1 | 106 | 0.710 | 0.0456 | Yes |
| 3 | Pdcd4 | 124 | 0.686 | 0.0667 | Yes |
| 4 | Gna15 | 271 | 0.588 | 0.0733 | Yes |
| 5 | Cdkn1b | 282 | 0.583 | 0.0916 | Yes |
| 6 | Gpx1 | 316 | 0.570 | 0.1074 | Yes |
| 7 | Wee1 | 329 | 0.564 | 0.1249 | Yes |
| 8 | Jun | 361 | 0.548 | 0.1402 | Yes |
| 9 | Mcl1 | 438 | 0.520 | 0.1507 | Yes |
| 10 | Brca1 | 668 | 0.450 | 0.1455 | Yes |
| 11 | Rnasel | 709 | 0.439 | 0.1565 | Yes |
| 12 | Cd69 | 931 | 0.394 | 0.1502 | Yes |
| 13 | Bcap31 | 963 | 0.390 | 0.1603 | Yes |
| 14 | Isg20 | 1088 | 0.371 | 0.1618 | Yes |
| 15 | Casp2 | 1167 | 0.358 | 0.1667 | Yes |
| 16 | Ppt1 | 1168 | 0.358 | 0.1785 | Yes |
| 17 | Psen1 | 1270 | 0.344 | 0.1810 | Yes |
| 18 | Dnm1l | 1383 | 0.329 | 0.1821 | Yes |
| 19 | Hmgb2 | 1462 | 0.320 | 0.1858 | Yes |
| 20 | Ifngr1 | 1531 | 0.312 | 0.1901 | Yes |
| 21 | Ifitm3 | 1534 | 0.312 | 0.2002 | Yes |
| 22 | Bcl2l2 | 1612 | 0.301 | 0.2034 | Yes |
| 23 | Etf1 | 1646 | 0.297 | 0.2103 | Yes |
| 24 | Igf2r | 2076 | 0.253 | 0.1813 | No |
| 25 | Hgf | 2101 | 0.251 | 0.1874 | No |
| 26 | Tgfbr3 | 2355 | 0.223 | 0.1727 | No |
| 27 | Mgmt | 2361 | 0.222 | 0.1796 | No |
| 28 | Irf1 | 2400 | 0.219 | 0.1835 | No |
| 29 | Sat1 | 2426 | 0.216 | 0.1884 | No |
| 30 | Gpx4 | 2470 | 0.212 | 0.1916 | No |
| 31 | Bcl10 | 2576 | 0.202 | 0.1891 | No |
| 32 | Lmna | 2591 | 0.201 | 0.1945 | No |
| 33 | Ier3 | 2627 | 0.197 | 0.1979 | No |
| 34 | Cd44 | 2654 | 0.195 | 0.2021 | No |
| 35 | Top2a | 2696 | 0.193 | 0.2049 | No |
| 36 | Timp2 | 2881 | 0.175 | 0.1946 | No |
| 37 | Rela | 2918 | 0.173 | 0.1971 | No |
| 38 | Cflar | 2955 | 0.169 | 0.1996 | No |
| 39 | Cdc25b | 3184 | 0.150 | 0.1847 | No |
| 40 | Bnip3l | 3345 | 0.137 | 0.1753 | No |
| 41 | Gch1 | 3350 | 0.136 | 0.1794 | No |
| 42 | Nefh | 3569 | 0.121 | 0.1644 | No |
| 43 | Casp4 | 3595 | 0.118 | 0.1661 | No |
| 44 | Tnfsf10 | 3732 | 0.109 | 0.1579 | No |
| 45 | Fdxr | 3972 | 0.091 | 0.1401 | No |
| 46 | App | 4121 | 0.079 | 0.1298 | No |
| 47 | Dffa | 4358 | 0.059 | 0.1112 | No |
| 48 | Diablo | 4420 | 0.055 | 0.1077 | No |
| 49 | H1f0 | 4544 | 0.046 | 0.0985 | No |
| 50 | Casp8 | 4625 | 0.040 | 0.0929 | No |
| 51 | Birc3 | 4815 | 0.026 | 0.0773 | No |
| 52 | Tap1 | 4874 | 0.020 | 0.0729 | No |
| 53 | Sqstm1 | 4937 | 0.016 | 0.0680 | No |
| 54 | F2r | 5037 | 0.008 | 0.0597 | No |
| 55 | Dnajc3 | 5059 | 0.007 | 0.0581 | No |
| 56 | Casp1 | 5135 | 0.002 | 0.0516 | No |
| 57 | Gadd45a | 5199 | -0.002 | 0.0462 | No |
| 58 | Casp6 | 5249 | -0.006 | 0.0421 | No |
| 59 | Rock1 | 5330 | -0.012 | 0.0356 | No |
| 60 | Cyld | 5377 | -0.015 | 0.0321 | No |
| 61 | Ank | 5627 | -0.035 | 0.0116 | No |
| 62 | Madd | 5642 | -0.037 | 0.0116 | No |
| 63 | Add1 | 5699 | -0.041 | 0.0080 | No |
| 64 | Plcb2 | 5704 | -0.041 | 0.0090 | No |
| 65 | Nedd9 | 5899 | -0.055 | -0.0061 | No |
| 66 | Bcl2l1 | 6122 | -0.072 | -0.0230 | No |
| 67 | Btg2 | 6123 | -0.072 | -0.0207 | No |
| 68 | Erbb3 | 6291 | -0.085 | -0.0324 | No |
| 69 | Ebp | 6414 | -0.096 | -0.0399 | No |
| 70 | Dap3 | 6597 | -0.110 | -0.0521 | No |
| 71 | Pdgfrb | 6630 | -0.112 | -0.0512 | No |
| 72 | Tspo | 6650 | -0.114 | -0.0491 | No |
| 73 | Smad7 | 6774 | -0.125 | -0.0557 | No |
| 74 | Txnip | 7233 | -0.161 | -0.0902 | No |
| 75 | Bcl2l11 | 7488 | -0.181 | -0.1064 | No |
| 76 | Casp3 | 7543 | -0.186 | -0.1050 | No |
| 77 | Sptan1 | 7578 | -0.189 | -0.1017 | No |
| 78 | Sod2 | 7587 | -0.190 | -0.0962 | No |
| 79 | Retsat | 7620 | -0.193 | -0.0926 | No |
| 80 | Cdk2 | 7729 | -0.202 | -0.0954 | No |
| 81 | Btg3 | 7748 | -0.204 | -0.0902 | No |
| 82 | Rhob | 7792 | -0.207 | -0.0872 | No |
| 83 | Cdkn1a | 7917 | -0.220 | -0.0907 | No |
| 84 | Casp7 | 7928 | -0.220 | -0.0844 | No |
| 85 | Slc20a1 | 8019 | -0.227 | -0.0847 | No |
| 86 | Ddit3 | 8327 | -0.251 | -0.1032 | No |
| 87 | Crebbp | 8420 | -0.261 | -0.1026 | No |
| 88 | Gsr | 8473 | -0.267 | -0.0984 | No |
| 89 | Dnaja1 | 8501 | -0.270 | -0.0918 | No |
| 90 | Pea15a | 8596 | -0.277 | -0.0909 | No |
| 91 | Dap | 8705 | -0.288 | -0.0909 | No |
| 92 | Pak1 | 8729 | -0.290 | -0.0833 | No |
| 93 | Tgfb2 | 8828 | -0.299 | -0.0820 | No |
| 94 | Ctnnb1 | 8911 | -0.306 | -0.0791 | No |
| 95 | Rara | 9312 | -0.347 | -0.1025 | No |
| 96 | Krt18 | 9483 | -0.367 | -0.1052 | No |
| 97 | Fas | 9567 | -0.374 | -0.1001 | No |
| 98 | Vdac2 | 9625 | -0.380 | -0.0926 | No |
| 99 | Ppp3r1 | 9896 | -0.412 | -0.1025 | No |
| 100 | Sod1 | 10034 | -0.432 | -0.1002 | No |
| 101 | Rhot2 | 10166 | -0.453 | -0.0968 | No |
| 102 | Sc5d | 10293 | -0.475 | -0.0921 | No |
| 103 | Psen2 | 10480 | -0.501 | -0.0919 | No |
| 104 | Satb1 | 10527 | -0.507 | -0.0792 | No |
| 105 | Gadd45b | 10548 | -0.510 | -0.0642 | No |
| 106 | Gsn | 10557 | -0.512 | -0.0481 | No |
| 107 | Xiap | 10679 | -0.534 | -0.0411 | No |
| 108 | Ptk2 | 10699 | -0.538 | -0.0250 | No |
| 109 | Bid | 11148 | -0.673 | -0.0419 | No |
| 110 | Casp9 | 11171 | -0.685 | -0.0213 | No |
| 111 | Ccnd2 | 11467 | -0.893 | -0.0176 | No |
| 112 | Bax | 11479 | -0.910 | 0.0113 | No |