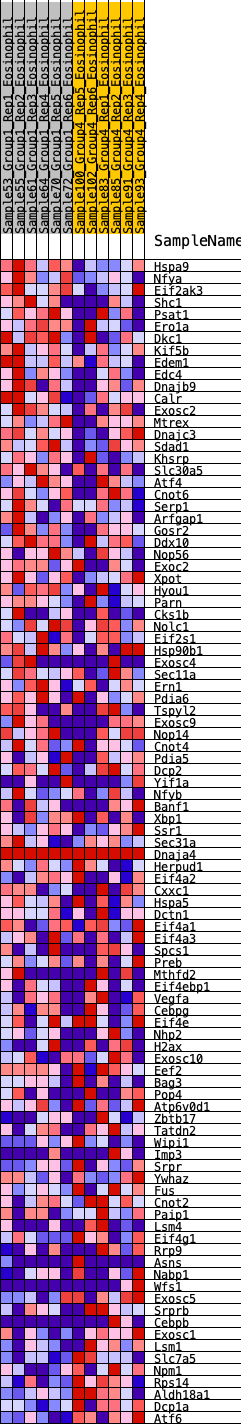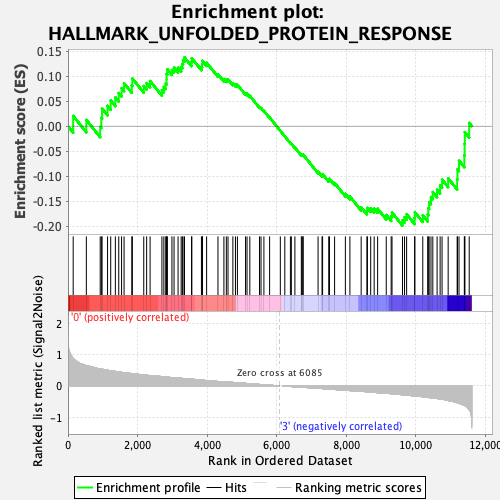
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.19875684 |
| Normalized Enrichment Score (NES) | -0.7565083 |
| Nominal p-value | 0.8360324 |
| FDR q-value | 0.8705284 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hspa9 | 149 | 0.876 | 0.0205 | No |
| 2 | Nfya | 527 | 0.649 | 0.0125 | No |
| 3 | Eif2ak3 | 926 | 0.544 | -0.0012 | No |
| 4 | Shc1 | 958 | 0.536 | 0.0165 | No |
| 5 | Psat1 | 980 | 0.533 | 0.0351 | No |
| 6 | Ero1a | 1137 | 0.499 | 0.0406 | No |
| 7 | Dkc1 | 1228 | 0.486 | 0.0513 | No |
| 8 | Kif5b | 1361 | 0.463 | 0.0576 | No |
| 9 | Edem1 | 1459 | 0.449 | 0.0663 | No |
| 10 | Edc4 | 1540 | 0.433 | 0.0759 | No |
| 11 | Dnajb9 | 1612 | 0.420 | 0.0858 | No |
| 12 | Calr | 1834 | 0.394 | 0.0816 | No |
| 13 | Exosc2 | 1850 | 0.391 | 0.0953 | No |
| 14 | Mtrex | 2178 | 0.352 | 0.0803 | No |
| 15 | Dnajc3 | 2261 | 0.341 | 0.0862 | No |
| 16 | Sdad1 | 2360 | 0.331 | 0.0903 | No |
| 17 | Khsrp | 2700 | 0.298 | 0.0723 | No |
| 18 | Slc30a5 | 2756 | 0.291 | 0.0786 | No |
| 19 | Atf4 | 2809 | 0.287 | 0.0850 | No |
| 20 | Cnot6 | 2834 | 0.284 | 0.0938 | No |
| 21 | Serp1 | 2835 | 0.284 | 0.1046 | No |
| 22 | Arfgap1 | 2857 | 0.282 | 0.1135 | No |
| 23 | Gosr2 | 2988 | 0.267 | 0.1124 | No |
| 24 | Ddx10 | 3049 | 0.260 | 0.1172 | No |
| 25 | Nop56 | 3164 | 0.250 | 0.1168 | No |
| 26 | Exoc2 | 3253 | 0.243 | 0.1185 | No |
| 27 | Xpot | 3288 | 0.239 | 0.1247 | No |
| 28 | Hyou1 | 3307 | 0.236 | 0.1321 | No |
| 29 | Parn | 3349 | 0.232 | 0.1374 | No |
| 30 | Cks1b | 3552 | 0.215 | 0.1281 | No |
| 31 | Nolc1 | 3560 | 0.214 | 0.1357 | No |
| 32 | Eif2s1 | 3836 | 0.186 | 0.1189 | No |
| 33 | Hsp90b1 | 3856 | 0.185 | 0.1243 | No |
| 34 | Exosc4 | 3866 | 0.184 | 0.1305 | No |
| 35 | Sec11a | 3985 | 0.171 | 0.1268 | No |
| 36 | Ern1 | 4311 | 0.141 | 0.1040 | No |
| 37 | Pdia6 | 4482 | 0.134 | 0.0943 | No |
| 38 | Tspyl2 | 4552 | 0.127 | 0.0932 | No |
| 39 | Exosc9 | 4600 | 0.123 | 0.0938 | No |
| 40 | Nop14 | 4740 | 0.111 | 0.0860 | No |
| 41 | Cnot4 | 4814 | 0.106 | 0.0837 | No |
| 42 | Pdia5 | 4870 | 0.101 | 0.0827 | No |
| 43 | Dcp2 | 5104 | 0.080 | 0.0656 | No |
| 44 | Yif1a | 5147 | 0.077 | 0.0649 | No |
| 45 | Nfyb | 5227 | 0.070 | 0.0607 | No |
| 46 | Banf1 | 5506 | 0.047 | 0.0383 | No |
| 47 | Xbp1 | 5549 | 0.043 | 0.0363 | No |
| 48 | Ssr1 | 5632 | 0.036 | 0.0305 | No |
| 49 | Sec31a | 5796 | 0.025 | 0.0173 | No |
| 50 | Dnaja4 | 6104 | 0.000 | -0.0093 | No |
| 51 | Herpud1 | 6233 | -0.004 | -0.0203 | No |
| 52 | Eif4a2 | 6394 | -0.017 | -0.0335 | No |
| 53 | Cxxc1 | 6421 | -0.019 | -0.0350 | No |
| 54 | Hspa5 | 6523 | -0.028 | -0.0428 | No |
| 55 | Dctn1 | 6708 | -0.037 | -0.0573 | No |
| 56 | Eif4a1 | 6726 | -0.039 | -0.0573 | No |
| 57 | Eif4a3 | 6750 | -0.041 | -0.0577 | No |
| 58 | Spcs1 | 6761 | -0.042 | -0.0570 | No |
| 59 | Preb | 7189 | -0.076 | -0.0912 | No |
| 60 | Mthfd2 | 7303 | -0.083 | -0.0978 | No |
| 61 | Eif4ebp1 | 7319 | -0.085 | -0.0959 | No |
| 62 | Vegfa | 7496 | -0.100 | -0.1073 | No |
| 63 | Cebpg | 7517 | -0.102 | -0.1052 | No |
| 64 | Eif4e | 7664 | -0.113 | -0.1136 | No |
| 65 | Nhp2 | 7974 | -0.139 | -0.1351 | No |
| 66 | H2ax | 8104 | -0.149 | -0.1406 | No |
| 67 | Exosc10 | 8428 | -0.170 | -0.1622 | No |
| 68 | Eef2 | 8593 | -0.186 | -0.1693 | No |
| 69 | Bag3 | 8608 | -0.187 | -0.1634 | No |
| 70 | Pop4 | 8702 | -0.197 | -0.1640 | No |
| 71 | Atp6v0d1 | 8802 | -0.206 | -0.1647 | No |
| 72 | Zbtb17 | 8901 | -0.215 | -0.1650 | No |
| 73 | Tatdn2 | 9149 | -0.234 | -0.1775 | No |
| 74 | Wipi1 | 9290 | -0.248 | -0.1802 | No |
| 75 | Imp3 | 9315 | -0.251 | -0.1727 | No |
| 76 | Srpr | 9616 | -0.282 | -0.1880 | Yes |
| 77 | Ywhaz | 9675 | -0.288 | -0.1820 | Yes |
| 78 | Fus | 9735 | -0.294 | -0.1759 | Yes |
| 79 | Cnot2 | 9965 | -0.319 | -0.1836 | Yes |
| 80 | Paip1 | 9972 | -0.320 | -0.1719 | Yes |
| 81 | Lsm4 | 10199 | -0.347 | -0.1783 | Yes |
| 82 | Eif4g1 | 10341 | -0.366 | -0.1766 | Yes |
| 83 | Rrp9 | 10358 | -0.369 | -0.1639 | Yes |
| 84 | Asns | 10382 | -0.373 | -0.1516 | Yes |
| 85 | Nabp1 | 10435 | -0.379 | -0.1417 | Yes |
| 86 | Wfs1 | 10488 | -0.382 | -0.1316 | Yes |
| 87 | Exosc5 | 10611 | -0.401 | -0.1269 | Yes |
| 88 | Srprb | 10698 | -0.418 | -0.1184 | Yes |
| 89 | Cebpb | 10754 | -0.427 | -0.1069 | Yes |
| 90 | Exosc1 | 10929 | -0.466 | -0.1042 | Yes |
| 91 | Lsm1 | 11188 | -0.535 | -0.1062 | Yes |
| 92 | Slc7a5 | 11197 | -0.539 | -0.0862 | Yes |
| 93 | Npm1 | 11241 | -0.553 | -0.0688 | Yes |
| 94 | Rps14 | 11394 | -0.618 | -0.0584 | Yes |
| 95 | Aldh18a1 | 11402 | -0.622 | -0.0353 | Yes |
| 96 | Dcp1a | 11409 | -0.628 | -0.0118 | Yes |
| 97 | Atf6 | 11534 | -0.762 | 0.0065 | Yes |