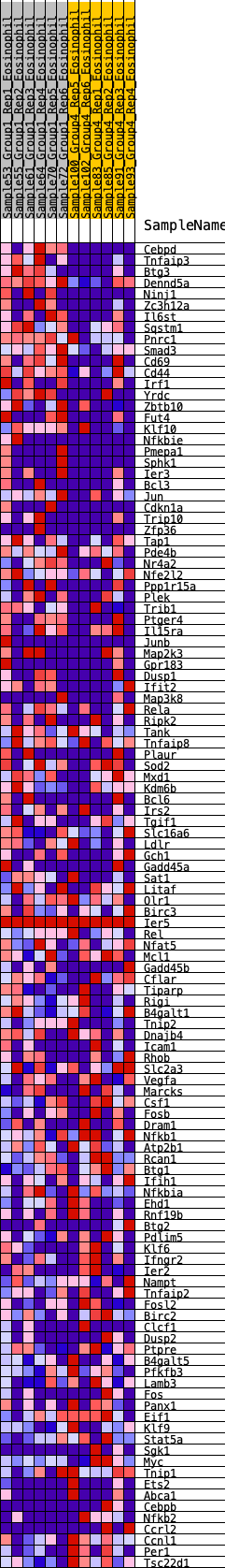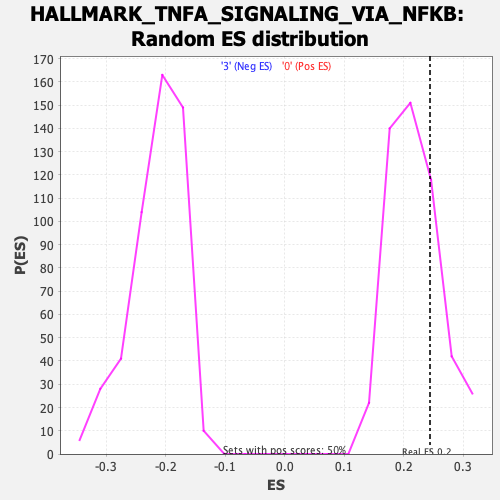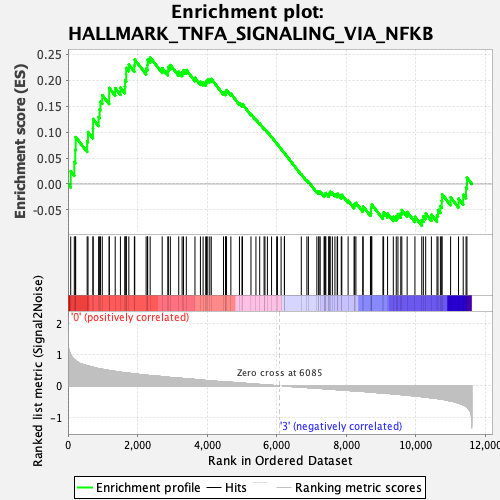
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.24406761 |
| Normalized Enrichment Score (NES) | 1.1211951 |
| Nominal p-value | 0.252505 |
| FDR q-value | 0.4356846 |
| FWER p-Value | 0.929 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cebpd | 78 | 1.001 | 0.0248 | Yes |
| 2 | Tnfaip3 | 181 | 0.839 | 0.0424 | Yes |
| 3 | Btg3 | 209 | 0.820 | 0.0659 | Yes |
| 4 | Dennd5a | 221 | 0.810 | 0.0905 | Yes |
| 5 | Ninj1 | 551 | 0.644 | 0.0822 | Yes |
| 6 | Zc3h12a | 575 | 0.636 | 0.1002 | Yes |
| 7 | Il6st | 717 | 0.599 | 0.1068 | Yes |
| 8 | Sqstm1 | 722 | 0.598 | 0.1254 | Yes |
| 9 | Pnrc1 | 880 | 0.552 | 0.1291 | Yes |
| 10 | Smad3 | 909 | 0.546 | 0.1439 | Yes |
| 11 | Cd69 | 933 | 0.542 | 0.1590 | Yes |
| 12 | Cd44 | 985 | 0.532 | 0.1713 | Yes |
| 13 | Irf1 | 1184 | 0.493 | 0.1696 | Yes |
| 14 | Yrdc | 1185 | 0.492 | 0.1852 | Yes |
| 15 | Zbtb10 | 1357 | 0.464 | 0.1849 | Yes |
| 16 | Fut4 | 1507 | 0.441 | 0.1859 | Yes |
| 17 | Klf10 | 1635 | 0.416 | 0.1879 | Yes |
| 18 | Nfkbie | 1643 | 0.416 | 0.2004 | Yes |
| 19 | Pmepa1 | 1674 | 0.412 | 0.2108 | Yes |
| 20 | Sphk1 | 1675 | 0.412 | 0.2238 | Yes |
| 21 | Ier3 | 1749 | 0.402 | 0.2302 | Yes |
| 22 | Bcl3 | 1908 | 0.385 | 0.2285 | Yes |
| 23 | Jun | 1919 | 0.383 | 0.2398 | Yes |
| 24 | Cdkn1a | 2246 | 0.343 | 0.2222 | Yes |
| 25 | Trip10 | 2285 | 0.337 | 0.2295 | Yes |
| 26 | Zfp36 | 2291 | 0.337 | 0.2397 | Yes |
| 27 | Tap1 | 2362 | 0.331 | 0.2441 | Yes |
| 28 | Pde4b | 2708 | 0.298 | 0.2234 | No |
| 29 | Nr4a2 | 2871 | 0.280 | 0.2182 | No |
| 30 | Nfe2l2 | 2884 | 0.278 | 0.2259 | No |
| 31 | Ppp1r15a | 2942 | 0.269 | 0.2294 | No |
| 32 | Plek | 3183 | 0.249 | 0.2163 | No |
| 33 | Trib1 | 3281 | 0.240 | 0.2155 | No |
| 34 | Ptger4 | 3321 | 0.235 | 0.2195 | No |
| 35 | Il15ra | 3401 | 0.227 | 0.2198 | No |
| 36 | Junb | 3650 | 0.206 | 0.2047 | No |
| 37 | Map2k3 | 3807 | 0.189 | 0.1971 | No |
| 38 | Gpr183 | 3884 | 0.182 | 0.1962 | No |
| 39 | Dusp1 | 3960 | 0.173 | 0.1951 | No |
| 40 | Ifit2 | 3981 | 0.172 | 0.1988 | No |
| 41 | Map3k8 | 4018 | 0.168 | 0.2010 | No |
| 42 | Rela | 4076 | 0.163 | 0.2011 | No |
| 43 | Ripk2 | 4119 | 0.160 | 0.2025 | No |
| 44 | Tank | 4470 | 0.136 | 0.1764 | No |
| 45 | Tnfaip8 | 4529 | 0.129 | 0.1754 | No |
| 46 | Plaur | 4538 | 0.128 | 0.1787 | No |
| 47 | Sod2 | 4558 | 0.127 | 0.1811 | No |
| 48 | Mxd1 | 4682 | 0.116 | 0.1741 | No |
| 49 | Kdm6b | 4929 | 0.095 | 0.1556 | No |
| 50 | Bcl6 | 5002 | 0.089 | 0.1522 | No |
| 51 | Irs2 | 5016 | 0.089 | 0.1539 | No |
| 52 | Tgif1 | 5260 | 0.067 | 0.1348 | No |
| 53 | Slc16a6 | 5403 | 0.055 | 0.1242 | No |
| 54 | Ldlr | 5511 | 0.046 | 0.1163 | No |
| 55 | Gch1 | 5637 | 0.036 | 0.1066 | No |
| 56 | Gadd45a | 5662 | 0.034 | 0.1056 | No |
| 57 | Sat1 | 5733 | 0.029 | 0.1004 | No |
| 58 | Litaf | 5852 | 0.020 | 0.0908 | No |
| 59 | Olr1 | 5999 | 0.007 | 0.0783 | No |
| 60 | Birc3 | 6021 | 0.005 | 0.0766 | No |
| 61 | Ier5 | 6126 | 0.000 | 0.0675 | No |
| 62 | Rel | 6220 | -0.003 | 0.0596 | No |
| 63 | Nfat5 | 6224 | -0.004 | 0.0594 | No |
| 64 | Mcl1 | 6707 | -0.037 | 0.0187 | No |
| 65 | Gadd45b | 6868 | -0.051 | 0.0063 | No |
| 66 | Cflar | 6911 | -0.055 | 0.0044 | No |
| 67 | Tiparp | 7154 | -0.072 | -0.0144 | No |
| 68 | Rigi | 7210 | -0.078 | -0.0167 | No |
| 69 | B4galt1 | 7212 | -0.078 | -0.0143 | No |
| 70 | Tnip2 | 7250 | -0.080 | -0.0150 | No |
| 71 | Dnajb4 | 7362 | -0.088 | -0.0219 | No |
| 72 | Icam1 | 7378 | -0.089 | -0.0204 | No |
| 73 | Rhob | 7395 | -0.091 | -0.0189 | No |
| 74 | Slc2a3 | 7412 | -0.092 | -0.0174 | No |
| 75 | Vegfa | 7496 | -0.100 | -0.0215 | No |
| 76 | Marcks | 7503 | -0.101 | -0.0188 | No |
| 77 | Csf1 | 7527 | -0.103 | -0.0176 | No |
| 78 | Fosb | 7536 | -0.104 | -0.0150 | No |
| 79 | Dram1 | 7600 | -0.109 | -0.0171 | No |
| 80 | Nfkb1 | 7674 | -0.114 | -0.0198 | No |
| 81 | Atp2b1 | 7738 | -0.119 | -0.0215 | No |
| 82 | Rcan1 | 7750 | -0.120 | -0.0187 | No |
| 83 | Btg1 | 7856 | -0.129 | -0.0238 | No |
| 84 | Ifih1 | 7872 | -0.131 | -0.0209 | No |
| 85 | Nfkbia | 8053 | -0.145 | -0.0320 | No |
| 86 | Ehd1 | 8224 | -0.154 | -0.0420 | No |
| 87 | Rnf19b | 8242 | -0.156 | -0.0385 | No |
| 88 | Btg2 | 8277 | -0.159 | -0.0365 | No |
| 89 | Pdlim5 | 8474 | -0.175 | -0.0480 | No |
| 90 | Klf6 | 8485 | -0.176 | -0.0433 | No |
| 91 | Ifngr2 | 8695 | -0.196 | -0.0553 | No |
| 92 | Ier2 | 8713 | -0.197 | -0.0506 | No |
| 93 | Nampt | 8715 | -0.198 | -0.0444 | No |
| 94 | Tnfaip2 | 8735 | -0.200 | -0.0398 | No |
| 95 | Fosl2 | 9054 | -0.225 | -0.0604 | No |
| 96 | Birc2 | 9073 | -0.227 | -0.0548 | No |
| 97 | Clcf1 | 9187 | -0.238 | -0.0571 | No |
| 98 | Dusp2 | 9352 | -0.255 | -0.0634 | No |
| 99 | Ptpre | 9437 | -0.263 | -0.0624 | No |
| 100 | B4galt5 | 9485 | -0.268 | -0.0580 | No |
| 101 | Pfkfb3 | 9567 | -0.276 | -0.0563 | No |
| 102 | Lamb3 | 9597 | -0.279 | -0.0501 | No |
| 103 | Fos | 9750 | -0.296 | -0.0540 | No |
| 104 | Panx1 | 9974 | -0.320 | -0.0633 | No |
| 105 | Eif1 | 10170 | -0.343 | -0.0694 | No |
| 106 | Klf9 | 10216 | -0.348 | -0.0624 | No |
| 107 | Stat5a | 10283 | -0.357 | -0.0569 | No |
| 108 | Sgk1 | 10447 | -0.380 | -0.0591 | No |
| 109 | Myc | 10608 | -0.401 | -0.0604 | No |
| 110 | Tnip1 | 10637 | -0.407 | -0.0500 | No |
| 111 | Ets2 | 10706 | -0.420 | -0.0426 | No |
| 112 | Abca1 | 10744 | -0.426 | -0.0324 | No |
| 113 | Cebpb | 10754 | -0.427 | -0.0197 | No |
| 114 | Nfkb2 | 10999 | -0.481 | -0.0258 | No |
| 115 | Ccrl2 | 11227 | -0.549 | -0.0282 | No |
| 116 | Ccnl1 | 11362 | -0.607 | -0.0208 | No |
| 117 | Per1 | 11443 | -0.654 | -0.0071 | No |
| 118 | Tsc22d1 | 11469 | -0.681 | 0.0122 | No |