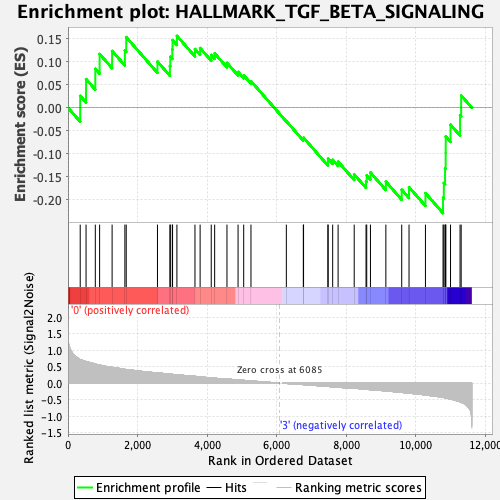
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
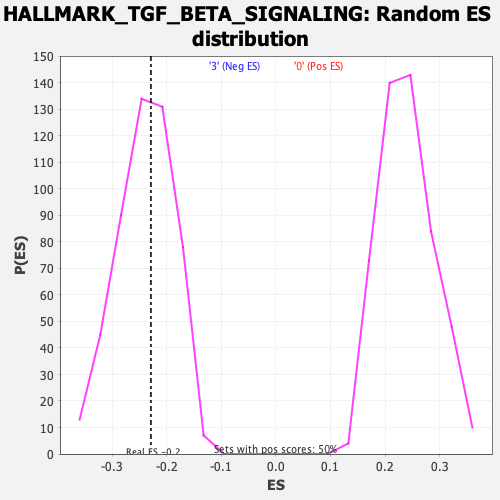
| Enrichment Score (ES) | -0.22942351 |
| Normalized Enrichment Score (NES) | -0.95517486 |
| Nominal p-value | 0.53815264 |
| FDR q-value | 0.9743047 |
| FWER p-Value | 0.986 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Smurf2 | 351 | 0.714 | 0.0256 | No |
| 2 | Skil | 519 | 0.651 | 0.0622 | No |
| 3 | Ctnnb1 | 785 | 0.578 | 0.0846 | No |
| 4 | Smad3 | 909 | 0.546 | 0.1168 | No |
| 5 | Ncor2 | 1268 | 0.478 | 0.1233 | No |
| 6 | Klf10 | 1635 | 0.416 | 0.1243 | No |
| 7 | Pmepa1 | 1674 | 0.412 | 0.1534 | No |
| 8 | Slc20a1 | 2572 | 0.312 | 0.1003 | No |
| 9 | Cdk9 | 2930 | 0.271 | 0.0906 | No |
| 10 | Ppp1r15a | 2942 | 0.269 | 0.1108 | No |
| 11 | Eng | 2999 | 0.266 | 0.1268 | No |
| 12 | Smurf1 | 3006 | 0.265 | 0.1471 | No |
| 13 | Map3k7 | 3131 | 0.253 | 0.1562 | No |
| 14 | Junb | 3650 | 0.206 | 0.1275 | No |
| 15 | Arid4b | 3798 | 0.190 | 0.1297 | No |
| 16 | Rhoa | 4118 | 0.160 | 0.1147 | No |
| 17 | Apc | 4216 | 0.151 | 0.1181 | No |
| 18 | Bmpr1a | 4570 | 0.126 | 0.0975 | No |
| 19 | Ppp1ca | 4889 | 0.099 | 0.0777 | No |
| 20 | Smad7 | 5051 | 0.084 | 0.0704 | No |
| 21 | Tgif1 | 5260 | 0.067 | 0.0577 | No |
| 22 | Ube2d3 | 6277 | -0.009 | -0.0294 | No |
| 23 | Hdac1 | 6765 | -0.042 | -0.0682 | No |
| 24 | Smad6 | 6772 | -0.043 | -0.0654 | No |
| 25 | Rab31 | 7473 | -0.098 | -0.1182 | No |
| 26 | Hipk2 | 7477 | -0.099 | -0.1107 | No |
| 27 | Furin | 7609 | -0.109 | -0.1135 | No |
| 28 | Ski | 7765 | -0.122 | -0.1174 | No |
| 29 | Fnta | 8228 | -0.154 | -0.1452 | No |
| 30 | Cdh1 | 8568 | -0.184 | -0.1601 | No |
| 31 | Trim33 | 8588 | -0.186 | -0.1472 | No |
| 32 | Ifngr2 | 8695 | -0.196 | -0.1410 | No |
| 33 | Acvr1 | 9138 | -0.233 | -0.1609 | No |
| 34 | Fkbp1a | 9594 | -0.279 | -0.1784 | No |
| 35 | Tgfb1 | 9804 | -0.301 | -0.1729 | No |
| 36 | Thbs1 | 10276 | -0.355 | -0.1858 | No |
| 37 | Sptbn1 | 10782 | -0.429 | -0.1958 | Yes |
| 38 | Bcar3 | 10805 | -0.434 | -0.1636 | Yes |
| 39 | Tjp1 | 10840 | -0.443 | -0.1319 | Yes |
| 40 | Ppm1a | 10858 | -0.450 | -0.0981 | Yes |
| 41 | Tgfbr1 | 10859 | -0.450 | -0.0628 | Yes |
| 42 | Smad1 | 10997 | -0.480 | -0.0370 | Yes |
| 43 | Xiap | 11271 | -0.566 | -0.0162 | Yes |
| 44 | Bmpr2 | 11303 | -0.579 | 0.0265 | Yes |