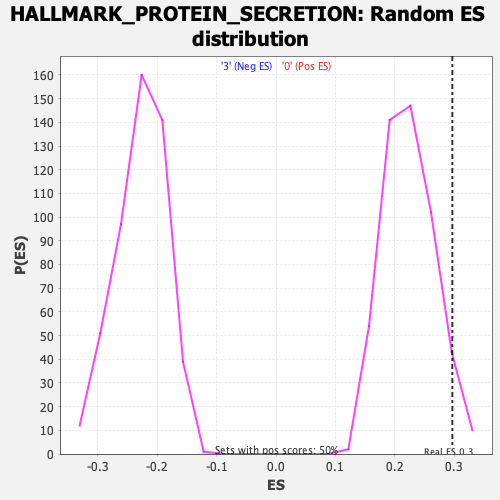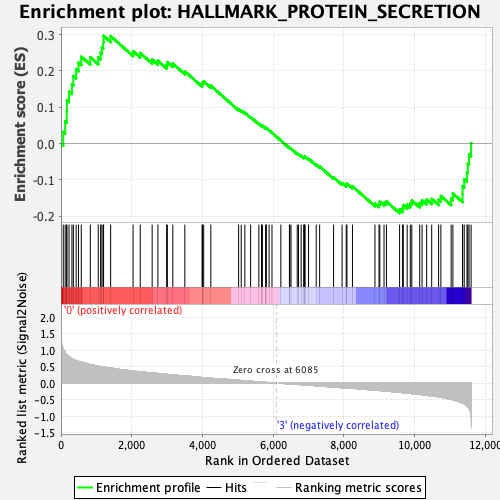
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.2966181 |
| Normalized Enrichment Score (NES) | 1.3292267 |
| Nominal p-value | 0.06212425 |
| FDR q-value | 0.24901663 |
| FWER p-Value | 0.6 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Scamp1 | 64 | 1.032 | 0.0320 | Yes |
| 2 | Yipf6 | 118 | 0.918 | 0.0609 | Yes |
| 3 | Anp32e | 161 | 0.863 | 0.0887 | Yes |
| 4 | Sod1 | 166 | 0.853 | 0.1195 | Yes |
| 5 | Tmx1 | 227 | 0.804 | 0.1435 | Yes |
| 6 | Atp7a | 309 | 0.743 | 0.1636 | Yes |
| 7 | Ctsc | 349 | 0.715 | 0.1862 | Yes |
| 8 | Snap23 | 428 | 0.688 | 0.2045 | Yes |
| 9 | Zw10 | 494 | 0.661 | 0.2230 | Yes |
| 10 | M6pr | 573 | 0.637 | 0.2394 | Yes |
| 11 | Krt18 | 829 | 0.563 | 0.2378 | Yes |
| 12 | Adam10 | 1053 | 0.518 | 0.2373 | Yes |
| 13 | Snx2 | 1120 | 0.501 | 0.2498 | Yes |
| 14 | Vamp3 | 1157 | 0.497 | 0.2648 | Yes |
| 15 | Napg | 1194 | 0.491 | 0.2796 | Yes |
| 16 | Copb2 | 1204 | 0.490 | 0.2966 | Yes |
| 17 | Arcn1 | 1404 | 0.455 | 0.2959 | No |
| 18 | Clta | 2038 | 0.367 | 0.2544 | No |
| 19 | Arfgef2 | 2241 | 0.343 | 0.2494 | No |
| 20 | Mon2 | 2578 | 0.311 | 0.2315 | No |
| 21 | Sgms1 | 2741 | 0.293 | 0.2281 | No |
| 22 | Gosr2 | 2988 | 0.267 | 0.2165 | No |
| 23 | Tpd52 | 3008 | 0.265 | 0.2245 | No |
| 24 | Lman1 | 3163 | 0.250 | 0.2203 | No |
| 25 | Gbf1 | 3502 | 0.219 | 0.1989 | No |
| 26 | Lamp2 | 3990 | 0.171 | 0.1629 | No |
| 27 | Ap1g1 | 4001 | 0.170 | 0.1682 | No |
| 28 | Rab5a | 4035 | 0.166 | 0.1714 | No |
| 29 | Sec24d | 4237 | 0.149 | 0.1594 | No |
| 30 | Golga4 | 5023 | 0.088 | 0.0945 | No |
| 31 | Dop1a | 5097 | 0.081 | 0.0911 | No |
| 32 | Tom1l1 | 5199 | 0.072 | 0.0850 | No |
| 33 | Cog2 | 5363 | 0.058 | 0.0729 | No |
| 34 | Ykt6 | 5596 | 0.039 | 0.0542 | No |
| 35 | Cltc | 5671 | 0.033 | 0.0490 | No |
| 36 | Ap2m1 | 5681 | 0.033 | 0.0494 | No |
| 37 | Dst | 5695 | 0.032 | 0.0494 | No |
| 38 | Rer1 | 5793 | 0.025 | 0.0419 | No |
| 39 | Sec31a | 5796 | 0.025 | 0.0427 | No |
| 40 | Vps4b | 5804 | 0.023 | 0.0429 | No |
| 41 | Mapk1 | 5885 | 0.017 | 0.0366 | No |
| 42 | Rab22a | 5967 | 0.009 | 0.0299 | No |
| 43 | Pam | 6218 | -0.003 | 0.0083 | No |
| 44 | Igf2r | 6460 | -0.022 | -0.0118 | No |
| 45 | Ap2b1 | 6472 | -0.023 | -0.0119 | No |
| 46 | Atp6v1h | 6505 | -0.026 | -0.0138 | No |
| 47 | Ap3b1 | 6681 | -0.035 | -0.0277 | No |
| 48 | Ergic3 | 6717 | -0.038 | -0.0293 | No |
| 49 | Arfgef1 | 6792 | -0.044 | -0.0342 | No |
| 50 | Stx7 | 6865 | -0.051 | -0.0386 | No |
| 51 | Ppt1 | 6878 | -0.052 | -0.0377 | No |
| 52 | Sec22b | 6888 | -0.053 | -0.0365 | No |
| 53 | Vps45 | 6903 | -0.054 | -0.0358 | No |
| 54 | Arfgap3 | 7001 | -0.061 | -0.0420 | No |
| 55 | Galc | 7218 | -0.078 | -0.0579 | No |
| 56 | Bet1 | 7315 | -0.085 | -0.0631 | No |
| 57 | Gnas | 7709 | -0.117 | -0.0930 | No |
| 58 | Rab14 | 7949 | -0.136 | -0.1087 | No |
| 59 | Tsg101 | 8066 | -0.145 | -0.1135 | No |
| 60 | Ocrl | 8087 | -0.147 | -0.1099 | No |
| 61 | Atp1a1 | 8243 | -0.156 | -0.1176 | No |
| 62 | Vamp4 | 8879 | -0.213 | -0.1650 | No |
| 63 | Cln5 | 8994 | -0.221 | -0.1668 | No |
| 64 | Scamp3 | 9016 | -0.223 | -0.1605 | No |
| 65 | Bnip3 | 9136 | -0.233 | -0.1624 | No |
| 66 | Cope | 9205 | -0.239 | -0.1595 | No |
| 67 | Stx12 | 9576 | -0.277 | -0.1816 | No |
| 68 | Copb1 | 9660 | -0.286 | -0.1783 | No |
| 69 | Kif1b | 9680 | -0.289 | -0.1694 | No |
| 70 | Ap2s1 | 9790 | -0.300 | -0.1680 | No |
| 71 | Arfip1 | 9879 | -0.310 | -0.1643 | No |
| 72 | Ap3s1 | 9921 | -0.315 | -0.1564 | No |
| 73 | Rab2a | 10145 | -0.339 | -0.1634 | No |
| 74 | Rab9 | 10212 | -0.347 | -0.1565 | No |
| 75 | Clcn3 | 10342 | -0.367 | -0.1543 | No |
| 76 | Rps6ka3 | 10484 | -0.382 | -0.1526 | No |
| 77 | Arf1 | 10679 | -0.414 | -0.1544 | No |
| 78 | Abca1 | 10744 | -0.426 | -0.1444 | No |
| 79 | Dnm1l | 11034 | -0.488 | -0.1517 | No |
| 80 | Stx16 | 11084 | -0.502 | -0.1377 | No |
| 81 | Ica1 | 11356 | -0.603 | -0.1392 | No |
| 82 | Napa | 11358 | -0.603 | -0.1173 | No |
| 83 | Gla | 11404 | -0.623 | -0.0985 | No |
| 84 | Uso1 | 11482 | -0.696 | -0.0799 | No |
| 85 | Tmed10 | 11506 | -0.724 | -0.0555 | No |
| 86 | Cd63 | 11540 | -0.768 | -0.0304 | No |
| 87 | Stam | 11598 | -0.995 | 0.0010 | No |