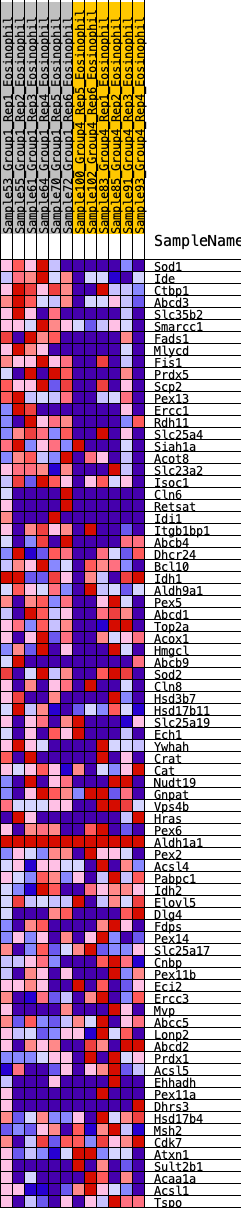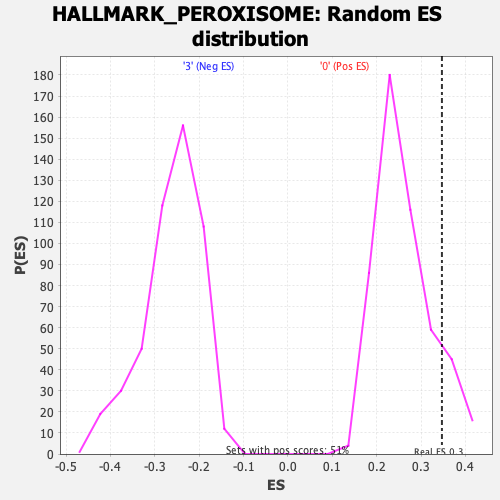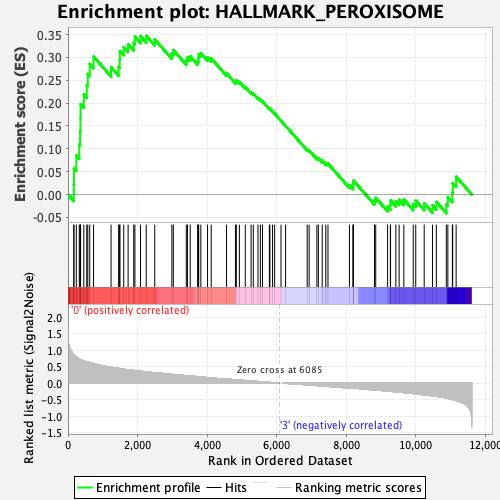
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.3470818 |
| Normalized Enrichment Score (NES) | 1.3380343 |
| Nominal p-value | 0.11857708 |
| FDR q-value | 0.26814803 |
| FWER p-Value | 0.593 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sod1 | 166 | 0.853 | 0.0220 | Yes |
| 2 | Ide | 171 | 0.849 | 0.0578 | Yes |
| 3 | Ctbp1 | 239 | 0.792 | 0.0858 | Yes |
| 4 | Abcd3 | 324 | 0.732 | 0.1097 | Yes |
| 5 | Slc35b2 | 347 | 0.717 | 0.1384 | Yes |
| 6 | Smarcc1 | 358 | 0.712 | 0.1679 | Yes |
| 7 | Fads1 | 362 | 0.710 | 0.1979 | Yes |
| 8 | Mlycd | 455 | 0.678 | 0.2189 | Yes |
| 9 | Fis1 | 539 | 0.646 | 0.2392 | Yes |
| 10 | Prdx5 | 569 | 0.638 | 0.2639 | Yes |
| 11 | Scp2 | 625 | 0.625 | 0.2858 | Yes |
| 12 | Pex13 | 735 | 0.595 | 0.3017 | Yes |
| 13 | Ercc1 | 1238 | 0.484 | 0.2788 | Yes |
| 14 | Rdh11 | 1455 | 0.449 | 0.2793 | Yes |
| 15 | Slc25a4 | 1485 | 0.445 | 0.2957 | Yes |
| 16 | Siah1a | 1492 | 0.443 | 0.3141 | Yes |
| 17 | Acot8 | 1602 | 0.422 | 0.3226 | Yes |
| 18 | Slc23a2 | 1728 | 0.406 | 0.3291 | Yes |
| 19 | Isoc1 | 1890 | 0.386 | 0.3316 | Yes |
| 20 | Cln6 | 1923 | 0.383 | 0.3452 | Yes |
| 21 | Retsat | 2084 | 0.361 | 0.3467 | Yes |
| 22 | Idi1 | 2249 | 0.343 | 0.3471 | Yes |
| 23 | Itgb1bp1 | 2494 | 0.317 | 0.3395 | No |
| 24 | Abcb4 | 2987 | 0.267 | 0.3082 | No |
| 25 | Dhcr24 | 3027 | 0.262 | 0.3160 | No |
| 26 | Bcl10 | 3400 | 0.227 | 0.2934 | No |
| 27 | Idh1 | 3434 | 0.223 | 0.3000 | No |
| 28 | Aldh9a1 | 3514 | 0.217 | 0.3025 | No |
| 29 | Pex5 | 3726 | 0.199 | 0.2926 | No |
| 30 | Abcd1 | 3753 | 0.196 | 0.2987 | No |
| 31 | Top2a | 3756 | 0.195 | 0.3069 | No |
| 32 | Acox1 | 3819 | 0.187 | 0.3095 | No |
| 33 | Hmgcl | 4012 | 0.168 | 0.3000 | No |
| 34 | Abcb9 | 4117 | 0.160 | 0.2978 | No |
| 35 | Sod2 | 4558 | 0.127 | 0.2651 | No |
| 36 | Cln8 | 4813 | 0.107 | 0.2476 | No |
| 37 | Hsd3b7 | 4839 | 0.104 | 0.2498 | No |
| 38 | Hsd17b11 | 4926 | 0.095 | 0.2465 | No |
| 39 | Slc25a19 | 5096 | 0.081 | 0.2353 | No |
| 40 | Ech1 | 5263 | 0.067 | 0.2237 | No |
| 41 | Ywhah | 5327 | 0.061 | 0.2208 | No |
| 42 | Crat | 5462 | 0.050 | 0.2114 | No |
| 43 | Cat | 5530 | 0.044 | 0.2075 | No |
| 44 | Nudt19 | 5592 | 0.039 | 0.2038 | No |
| 45 | Gnpat | 5788 | 0.025 | 0.1880 | No |
| 46 | Vps4b | 5804 | 0.023 | 0.1877 | No |
| 47 | Hras | 5881 | 0.017 | 0.1818 | No |
| 48 | Pex6 | 5941 | 0.011 | 0.1772 | No |
| 49 | Aldh1a1 | 6124 | 0.000 | 0.1614 | No |
| 50 | Pex2 | 6255 | -0.006 | 0.1504 | No |
| 51 | Acsl4 | 6876 | -0.052 | 0.0989 | No |
| 52 | Pabpc1 | 6932 | -0.056 | 0.0965 | No |
| 53 | Idh2 | 7155 | -0.072 | 0.0803 | No |
| 54 | Elovl5 | 7197 | -0.076 | 0.0800 | No |
| 55 | Dlg4 | 7308 | -0.084 | 0.0741 | No |
| 56 | Fdps | 7411 | -0.092 | 0.0692 | No |
| 57 | Pex14 | 7474 | -0.099 | 0.0680 | No |
| 58 | Slc25a17 | 8091 | -0.148 | 0.0208 | No |
| 59 | Cnbp | 8194 | -0.151 | 0.0185 | No |
| 60 | Pex11b | 8199 | -0.151 | 0.0246 | No |
| 61 | Eci2 | 8205 | -0.152 | 0.0306 | No |
| 62 | Ercc3 | 8811 | -0.207 | -0.0130 | No |
| 63 | Mvp | 8847 | -0.210 | -0.0071 | No |
| 64 | Abcc5 | 9188 | -0.238 | -0.0264 | No |
| 65 | Lonp2 | 9268 | -0.245 | -0.0228 | No |
| 66 | Abcd2 | 9273 | -0.246 | -0.0127 | No |
| 67 | Prdx1 | 9425 | -0.263 | -0.0146 | No |
| 68 | Acsl5 | 9519 | -0.272 | -0.0111 | No |
| 69 | Ehhadh | 9655 | -0.285 | -0.0106 | No |
| 70 | Pex11a | 9925 | -0.315 | -0.0205 | No |
| 71 | Dhrs3 | 9997 | -0.323 | -0.0129 | No |
| 72 | Hsd17b4 | 10240 | -0.352 | -0.0189 | No |
| 73 | Msh2 | 10480 | -0.381 | -0.0233 | No |
| 74 | Cdk7 | 10589 | -0.396 | -0.0158 | No |
| 75 | Atxn1 | 10875 | -0.455 | -0.0211 | No |
| 76 | Sult2b1 | 10920 | -0.464 | -0.0051 | No |
| 77 | Acaa1a | 11050 | -0.493 | 0.0047 | No |
| 78 | Acsl1 | 11060 | -0.496 | 0.0251 | No |
| 79 | Tspo | 11158 | -0.527 | 0.0391 | No |