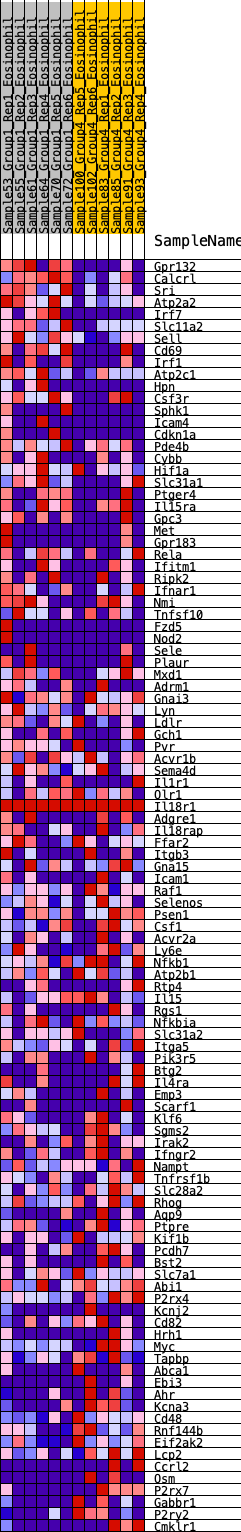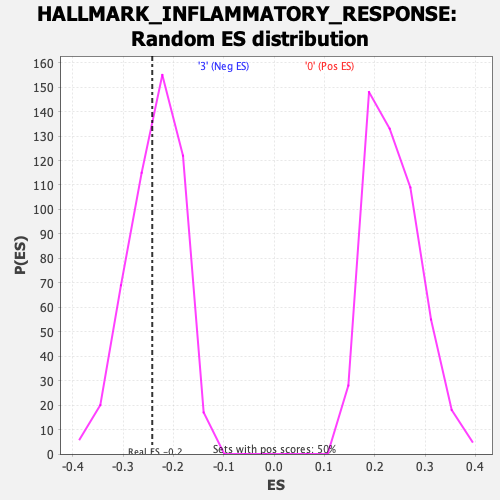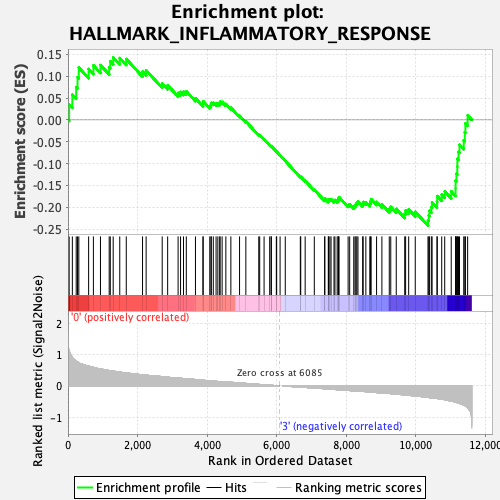
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.24203686 |
| Normalized Enrichment Score (NES) | -1.026897 |
| Nominal p-value | 0.41666666 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.963 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpr132 | 33 | 1.129 | 0.0352 | No |
| 2 | Calcrl | 125 | 0.907 | 0.0579 | No |
| 3 | Sri | 236 | 0.796 | 0.0752 | No |
| 4 | Atp2a2 | 274 | 0.765 | 0.0978 | No |
| 5 | Irf7 | 311 | 0.742 | 0.1197 | No |
| 6 | Slc11a2 | 593 | 0.632 | 0.1166 | No |
| 7 | Sell | 730 | 0.596 | 0.1249 | No |
| 8 | Cd69 | 933 | 0.542 | 0.1256 | No |
| 9 | Irf1 | 1184 | 0.493 | 0.1205 | No |
| 10 | Atp2c1 | 1216 | 0.487 | 0.1342 | No |
| 11 | Hpn | 1299 | 0.473 | 0.1431 | No |
| 12 | Csf3r | 1488 | 0.444 | 0.1417 | No |
| 13 | Sphk1 | 1675 | 0.412 | 0.1395 | No |
| 14 | Icam4 | 2143 | 0.354 | 0.1108 | No |
| 15 | Cdkn1a | 2246 | 0.343 | 0.1135 | No |
| 16 | Pde4b | 2708 | 0.298 | 0.0835 | No |
| 17 | Cybb | 2865 | 0.281 | 0.0794 | No |
| 18 | Hif1a | 3165 | 0.250 | 0.0618 | No |
| 19 | Slc31a1 | 3236 | 0.245 | 0.0640 | No |
| 20 | Ptger4 | 3321 | 0.235 | 0.0646 | No |
| 21 | Il15ra | 3401 | 0.227 | 0.0654 | No |
| 22 | Gpc3 | 3664 | 0.206 | 0.0496 | No |
| 23 | Met | 3881 | 0.182 | 0.0369 | No |
| 24 | Gpr183 | 3884 | 0.182 | 0.0429 | No |
| 25 | Rela | 4076 | 0.163 | 0.0318 | No |
| 26 | Ifitm1 | 4111 | 0.161 | 0.0342 | No |
| 27 | Ripk2 | 4119 | 0.160 | 0.0390 | No |
| 28 | Ifnar1 | 4177 | 0.155 | 0.0393 | No |
| 29 | Nmi | 4257 | 0.147 | 0.0374 | No |
| 30 | Tnfsf10 | 4308 | 0.141 | 0.0378 | No |
| 31 | Fzd5 | 4362 | 0.138 | 0.0379 | No |
| 32 | Nod2 | 4372 | 0.138 | 0.0417 | No |
| 33 | Sele | 4431 | 0.137 | 0.0413 | No |
| 34 | Plaur | 4538 | 0.128 | 0.0364 | No |
| 35 | Mxd1 | 4682 | 0.116 | 0.0279 | No |
| 36 | Adrm1 | 4932 | 0.095 | 0.0095 | No |
| 37 | Gnai3 | 5113 | 0.079 | -0.0035 | No |
| 38 | Lyn | 5484 | 0.048 | -0.0340 | No |
| 39 | Ldlr | 5511 | 0.046 | -0.0347 | No |
| 40 | Gch1 | 5637 | 0.036 | -0.0444 | No |
| 41 | Pvr | 5798 | 0.024 | -0.0575 | No |
| 42 | Acvr1b | 5838 | 0.020 | -0.0602 | No |
| 43 | Sema4d | 5847 | 0.020 | -0.0602 | No |
| 44 | Il1r1 | 5989 | 0.007 | -0.0722 | No |
| 45 | Olr1 | 5999 | 0.007 | -0.0728 | No |
| 46 | Il18r1 | 6097 | 0.000 | -0.0812 | No |
| 47 | Adgre1 | 6246 | -0.006 | -0.0939 | No |
| 48 | Il18rap | 6679 | -0.034 | -0.1303 | No |
| 49 | Ffar2 | 6691 | -0.035 | -0.1300 | No |
| 50 | Itgb3 | 6818 | -0.046 | -0.1394 | No |
| 51 | Gna15 | 7081 | -0.067 | -0.1599 | No |
| 52 | Icam1 | 7378 | -0.089 | -0.1827 | No |
| 53 | Raf1 | 7383 | -0.090 | -0.1800 | No |
| 54 | Selenos | 7485 | -0.100 | -0.1854 | No |
| 55 | Psen1 | 7487 | -0.100 | -0.1821 | No |
| 56 | Csf1 | 7527 | -0.103 | -0.1820 | No |
| 57 | Acvr2a | 7562 | -0.106 | -0.1814 | No |
| 58 | Ly6e | 7647 | -0.112 | -0.1850 | No |
| 59 | Nfkb1 | 7674 | -0.114 | -0.1834 | No |
| 60 | Atp2b1 | 7738 | -0.119 | -0.1848 | No |
| 61 | Rtp4 | 7770 | -0.122 | -0.1834 | No |
| 62 | Il15 | 7775 | -0.122 | -0.1796 | No |
| 63 | Rgs1 | 7793 | -0.124 | -0.1769 | No |
| 64 | Nfkbia | 8053 | -0.145 | -0.1946 | No |
| 65 | Slc31a2 | 8098 | -0.148 | -0.1934 | No |
| 66 | Itga5 | 8215 | -0.153 | -0.1983 | No |
| 67 | Pik3r5 | 8251 | -0.157 | -0.1961 | No |
| 68 | Btg2 | 8277 | -0.159 | -0.1929 | No |
| 69 | Il4ra | 8304 | -0.161 | -0.1897 | No |
| 70 | Emp3 | 8336 | -0.161 | -0.1869 | No |
| 71 | Scarf1 | 8471 | -0.174 | -0.1927 | No |
| 72 | Klf6 | 8485 | -0.176 | -0.1879 | No |
| 73 | Sgms2 | 8563 | -0.183 | -0.1884 | No |
| 74 | Irak2 | 8677 | -0.195 | -0.1917 | No |
| 75 | Ifngr2 | 8695 | -0.196 | -0.1865 | No |
| 76 | Nampt | 8715 | -0.198 | -0.1815 | No |
| 77 | Tnfrsf1b | 8871 | -0.212 | -0.1878 | No |
| 78 | Slc28a2 | 9024 | -0.224 | -0.1935 | No |
| 79 | Rhog | 9236 | -0.242 | -0.2037 | No |
| 80 | Aqp9 | 9279 | -0.246 | -0.1990 | No |
| 81 | Ptpre | 9437 | -0.263 | -0.2038 | No |
| 82 | Kif1b | 9680 | -0.289 | -0.2151 | No |
| 83 | Pcdh7 | 9706 | -0.292 | -0.2074 | No |
| 84 | Bst2 | 9794 | -0.301 | -0.2048 | No |
| 85 | Slc7a1 | 9983 | -0.322 | -0.2103 | No |
| 86 | Abi1 | 10349 | -0.367 | -0.2297 | Yes |
| 87 | P2rx4 | 10372 | -0.371 | -0.2191 | Yes |
| 88 | Kcnj2 | 10390 | -0.373 | -0.2079 | Yes |
| 89 | Cd82 | 10445 | -0.380 | -0.1998 | Yes |
| 90 | Hrh1 | 10467 | -0.381 | -0.1888 | Yes |
| 91 | Myc | 10608 | -0.401 | -0.1874 | Yes |
| 92 | Tapbp | 10615 | -0.402 | -0.1744 | Yes |
| 93 | Abca1 | 10744 | -0.426 | -0.1712 | Yes |
| 94 | Ebi3 | 10830 | -0.441 | -0.1637 | Yes |
| 95 | Ahr | 11017 | -0.485 | -0.1635 | Yes |
| 96 | Kcna3 | 11142 | -0.523 | -0.1566 | Yes |
| 97 | Cd48 | 11143 | -0.523 | -0.1390 | Yes |
| 98 | Rnf144b | 11167 | -0.529 | -0.1232 | Yes |
| 99 | Eif2ak2 | 11192 | -0.537 | -0.1071 | Yes |
| 100 | Lcp2 | 11194 | -0.538 | -0.0891 | Yes |
| 101 | Ccrl2 | 11227 | -0.549 | -0.0733 | Yes |
| 102 | Osm | 11255 | -0.558 | -0.0568 | Yes |
| 103 | P2rx7 | 11377 | -0.614 | -0.0466 | Yes |
| 104 | Gabbr1 | 11408 | -0.626 | -0.0282 | Yes |
| 105 | P2ry2 | 11425 | -0.640 | -0.0079 | Yes |
| 106 | Cmklr1 | 11489 | -0.707 | 0.0104 | Yes |