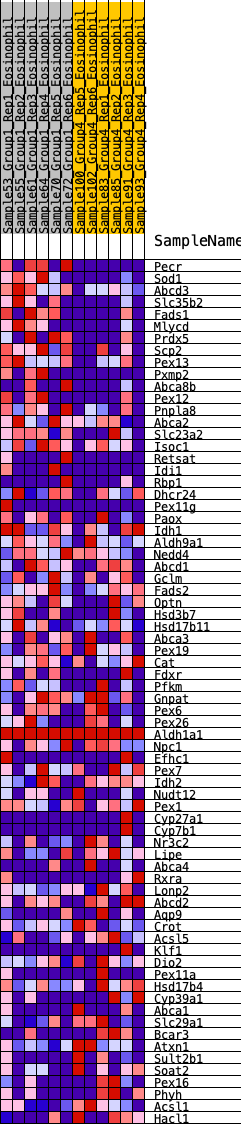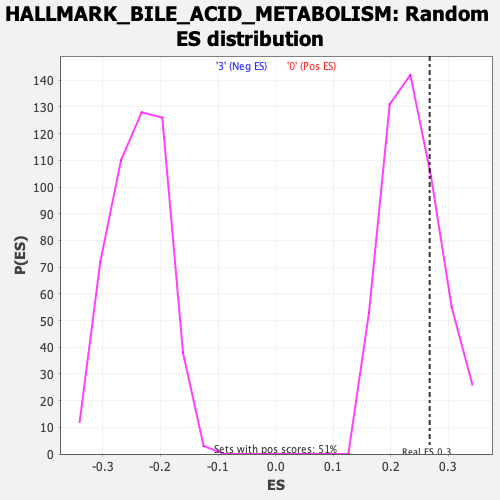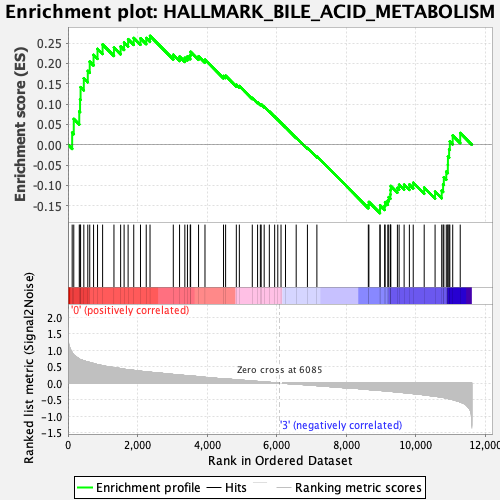
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.26792628 |
| Normalized Enrichment Score (NES) | 1.1286545 |
| Nominal p-value | 0.25244617 |
| FDR q-value | 0.46641213 |
| FWER p-Value | 0.923 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pecr | 120 | 0.915 | 0.0298 | Yes |
| 2 | Sod1 | 166 | 0.853 | 0.0635 | Yes |
| 3 | Abcd3 | 324 | 0.732 | 0.0820 | Yes |
| 4 | Slc35b2 | 347 | 0.717 | 0.1116 | Yes |
| 5 | Fads1 | 362 | 0.710 | 0.1416 | Yes |
| 6 | Mlycd | 455 | 0.678 | 0.1635 | Yes |
| 7 | Prdx5 | 569 | 0.638 | 0.1817 | Yes |
| 8 | Scp2 | 625 | 0.625 | 0.2044 | Yes |
| 9 | Pex13 | 735 | 0.595 | 0.2211 | Yes |
| 10 | Pxmp2 | 849 | 0.558 | 0.2359 | Yes |
| 11 | Abca8b | 995 | 0.530 | 0.2466 | Yes |
| 12 | Pex12 | 1321 | 0.468 | 0.2390 | Yes |
| 13 | Pnpla8 | 1513 | 0.440 | 0.2418 | Yes |
| 14 | Abca2 | 1613 | 0.420 | 0.2517 | Yes |
| 15 | Slc23a2 | 1728 | 0.406 | 0.2597 | Yes |
| 16 | Isoc1 | 1890 | 0.386 | 0.2627 | Yes |
| 17 | Retsat | 2084 | 0.361 | 0.2619 | Yes |
| 18 | Idi1 | 2249 | 0.343 | 0.2627 | Yes |
| 19 | Rbp1 | 2358 | 0.331 | 0.2679 | Yes |
| 20 | Dhcr24 | 3027 | 0.262 | 0.2216 | No |
| 21 | Pex11g | 3207 | 0.247 | 0.2169 | No |
| 22 | Paox | 3359 | 0.231 | 0.2140 | No |
| 23 | Idh1 | 3434 | 0.223 | 0.2173 | No |
| 24 | Aldh9a1 | 3514 | 0.217 | 0.2200 | No |
| 25 | Nedd4 | 3525 | 0.216 | 0.2287 | No |
| 26 | Abcd1 | 3753 | 0.196 | 0.2176 | No |
| 27 | Gclm | 3938 | 0.176 | 0.2094 | No |
| 28 | Fads2 | 4471 | 0.136 | 0.1693 | No |
| 29 | Optn | 4531 | 0.129 | 0.1698 | No |
| 30 | Hsd3b7 | 4839 | 0.104 | 0.1478 | No |
| 31 | Hsd17b11 | 4926 | 0.095 | 0.1445 | No |
| 32 | Abca3 | 5298 | 0.063 | 0.1151 | No |
| 33 | Pex19 | 5450 | 0.051 | 0.1043 | No |
| 34 | Cat | 5530 | 0.044 | 0.0994 | No |
| 35 | Fdxr | 5553 | 0.043 | 0.0994 | No |
| 36 | Pfkm | 5640 | 0.036 | 0.0935 | No |
| 37 | Gnpat | 5788 | 0.025 | 0.0819 | No |
| 38 | Pex6 | 5941 | 0.011 | 0.0692 | No |
| 39 | Pex26 | 6031 | 0.004 | 0.0616 | No |
| 40 | Aldh1a1 | 6124 | 0.000 | 0.0537 | No |
| 41 | Npc1 | 6253 | -0.006 | 0.0429 | No |
| 42 | Efhc1 | 6560 | -0.029 | 0.0176 | No |
| 43 | Pex7 | 6883 | -0.053 | -0.0080 | No |
| 44 | Idh2 | 7155 | -0.072 | -0.0283 | No |
| 45 | Nudt12 | 8634 | -0.190 | -0.1481 | No |
| 46 | Pex1 | 8650 | -0.192 | -0.1409 | No |
| 47 | Cyp27a1 | 8969 | -0.220 | -0.1588 | No |
| 48 | Cyp7b1 | 8972 | -0.220 | -0.1493 | No |
| 49 | Nr3c2 | 9104 | -0.230 | -0.1505 | No |
| 50 | Lipe | 9116 | -0.231 | -0.1413 | No |
| 51 | Abca4 | 9194 | -0.239 | -0.1375 | No |
| 52 | Rxra | 9220 | -0.241 | -0.1291 | No |
| 53 | Lonp2 | 9268 | -0.245 | -0.1224 | No |
| 54 | Abcd2 | 9273 | -0.246 | -0.1119 | No |
| 55 | Aqp9 | 9279 | -0.246 | -0.1016 | No |
| 56 | Crot | 9470 | -0.267 | -0.1063 | No |
| 57 | Acsl5 | 9519 | -0.272 | -0.0985 | No |
| 58 | Klf1 | 9663 | -0.286 | -0.0983 | No |
| 59 | Dio2 | 9814 | -0.303 | -0.0980 | No |
| 60 | Pex11a | 9925 | -0.315 | -0.0937 | No |
| 61 | Hsd17b4 | 10240 | -0.352 | -0.1054 | No |
| 62 | Cyp39a1 | 10553 | -0.389 | -0.1154 | No |
| 63 | Abca1 | 10744 | -0.426 | -0.1131 | No |
| 64 | Slc29a1 | 10784 | -0.429 | -0.0976 | No |
| 65 | Bcar3 | 10805 | -0.434 | -0.0802 | No |
| 66 | Atxn1 | 10875 | -0.455 | -0.0662 | No |
| 67 | Sult2b1 | 10920 | -0.464 | -0.0496 | No |
| 68 | Soat2 | 10923 | -0.465 | -0.0293 | No |
| 69 | Pex16 | 10957 | -0.472 | -0.0114 | No |
| 70 | Phyh | 10977 | -0.476 | 0.0079 | No |
| 71 | Acsl1 | 11060 | -0.496 | 0.0226 | No |
| 72 | Hacl1 | 11276 | -0.567 | 0.0289 | No |