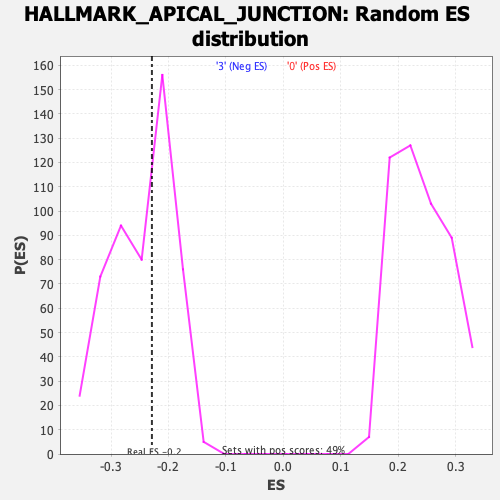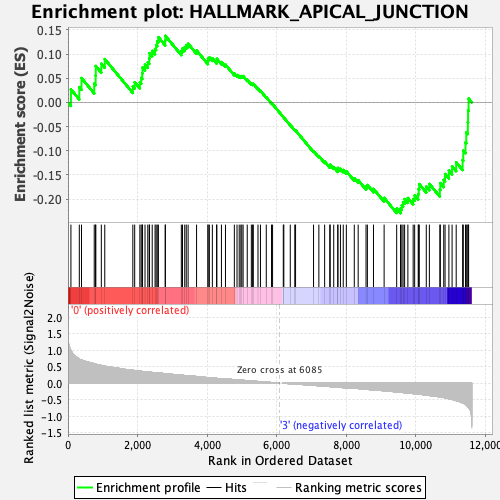
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group4.Eosinophil_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | -0.228863 |
| Normalized Enrichment Score (NES) | -0.9328008 |
| Nominal p-value | 0.53149605 |
| FDR q-value | 0.79804265 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tspan4 | 84 | 0.990 | 0.0271 | No |
| 2 | Pard6g | 325 | 0.730 | 0.0316 | No |
| 3 | Lima1 | 388 | 0.700 | 0.0505 | No |
| 4 | Sympk | 754 | 0.587 | 0.0391 | No |
| 5 | Pals1 | 794 | 0.576 | 0.0557 | No |
| 6 | Exoc4 | 798 | 0.574 | 0.0754 | No |
| 7 | Shc1 | 958 | 0.536 | 0.0802 | No |
| 8 | Vav2 | 1057 | 0.516 | 0.0896 | No |
| 9 | Mpzl1 | 1864 | 0.389 | 0.0331 | No |
| 10 | Tial1 | 1917 | 0.385 | 0.0419 | No |
| 11 | Taok2 | 2066 | 0.363 | 0.0416 | No |
| 12 | Skap2 | 2104 | 0.359 | 0.0509 | No |
| 13 | Actg1 | 2135 | 0.354 | 0.0606 | No |
| 14 | Icam4 | 2143 | 0.354 | 0.0722 | No |
| 15 | Itga9 | 2214 | 0.347 | 0.0782 | No |
| 16 | Nrap | 2294 | 0.337 | 0.0830 | No |
| 17 | Pkd1 | 2335 | 0.333 | 0.0911 | No |
| 18 | Map3k20 | 2344 | 0.332 | 0.1019 | No |
| 19 | Dhx16 | 2421 | 0.324 | 0.1066 | No |
| 20 | Pten | 2503 | 0.317 | 0.1105 | No |
| 21 | Amigo1 | 2533 | 0.315 | 0.1189 | No |
| 22 | Epb41l2 | 2566 | 0.313 | 0.1270 | No |
| 23 | Adam15 | 2599 | 0.308 | 0.1349 | No |
| 24 | Amigo2 | 2795 | 0.287 | 0.1280 | No |
| 25 | Cadm2 | 2799 | 0.287 | 0.1376 | No |
| 26 | Gtf2f1 | 3258 | 0.243 | 0.1063 | No |
| 27 | Ikbkg | 3292 | 0.238 | 0.1117 | No |
| 28 | Myh9 | 3356 | 0.231 | 0.1142 | No |
| 29 | Pbx2 | 3399 | 0.227 | 0.1185 | No |
| 30 | Vwf | 3453 | 0.221 | 0.1215 | No |
| 31 | Insig1 | 3696 | 0.202 | 0.1075 | No |
| 32 | Wasl | 4024 | 0.168 | 0.0849 | No |
| 33 | Ctnna1 | 4025 | 0.167 | 0.0907 | No |
| 34 | Zyx | 4064 | 0.164 | 0.0931 | No |
| 35 | Layn | 4150 | 0.158 | 0.0912 | No |
| 36 | Actn4 | 4273 | 0.145 | 0.0856 | No |
| 37 | Cdk8 | 4278 | 0.145 | 0.0903 | No |
| 38 | Jup | 4411 | 0.137 | 0.0836 | No |
| 39 | Ctnnd1 | 4527 | 0.129 | 0.0781 | No |
| 40 | Nlgn2 | 4782 | 0.109 | 0.0598 | No |
| 41 | Nf2 | 4859 | 0.102 | 0.0568 | No |
| 42 | Ptprc | 4922 | 0.096 | 0.0547 | No |
| 43 | Stx4a | 4969 | 0.092 | 0.0539 | No |
| 44 | Cnn2 | 4998 | 0.089 | 0.0546 | No |
| 45 | Arpc2 | 5034 | 0.087 | 0.0545 | No |
| 46 | Akt2 | 5153 | 0.076 | 0.0469 | No |
| 47 | Nf1 | 5271 | 0.066 | 0.0390 | No |
| 48 | Itgb1 | 5307 | 0.062 | 0.0381 | No |
| 49 | Ywhah | 5327 | 0.061 | 0.0386 | No |
| 50 | Crat | 5462 | 0.050 | 0.0287 | No |
| 51 | Tmem8b | 5533 | 0.044 | 0.0241 | No |
| 52 | Vasp | 5700 | 0.031 | 0.0108 | No |
| 53 | Icam2 | 5851 | 0.020 | -0.0015 | No |
| 54 | Hras | 5881 | 0.017 | -0.0035 | No |
| 55 | Vcam1 | 6189 | -0.001 | -0.0301 | No |
| 56 | Atp1a3 | 6205 | -0.002 | -0.0313 | No |
| 57 | Cd274 | 6390 | -0.017 | -0.0467 | No |
| 58 | Myh10 | 6520 | -0.028 | -0.0570 | No |
| 59 | Cldn15 | 6542 | -0.029 | -0.0578 | No |
| 60 | Tsc1 | 7059 | -0.065 | -0.1004 | No |
| 61 | B4galt1 | 7212 | -0.078 | -0.1109 | No |
| 62 | Icam1 | 7378 | -0.089 | -0.1221 | No |
| 63 | Plcg1 | 7526 | -0.103 | -0.1313 | No |
| 64 | Hadh | 7537 | -0.104 | -0.1286 | No |
| 65 | Rasa1 | 7639 | -0.111 | -0.1335 | No |
| 66 | Cd34 | 7757 | -0.121 | -0.1395 | No |
| 67 | Map4k2 | 7758 | -0.121 | -0.1353 | No |
| 68 | Bmp1 | 7832 | -0.127 | -0.1372 | No |
| 69 | Nectin1 | 7913 | -0.134 | -0.1395 | No |
| 70 | Baiap2 | 8002 | -0.141 | -0.1423 | No |
| 71 | Cap1 | 8230 | -0.155 | -0.1567 | No |
| 72 | Myl12b | 8342 | -0.162 | -0.1607 | No |
| 73 | Cdh1 | 8568 | -0.184 | -0.1739 | No |
| 74 | Tgfbi | 8607 | -0.187 | -0.1707 | No |
| 75 | Rac2 | 8782 | -0.204 | -0.1787 | No |
| 76 | Ptk2 | 9088 | -0.229 | -0.1973 | No |
| 77 | Arhgef6 | 9449 | -0.264 | -0.2194 | Yes |
| 78 | Itga10 | 9559 | -0.275 | -0.2193 | Yes |
| 79 | Lamb3 | 9597 | -0.279 | -0.2128 | Yes |
| 80 | Pfn1 | 9633 | -0.283 | -0.2060 | Yes |
| 81 | Akt3 | 9677 | -0.289 | -0.1997 | Yes |
| 82 | Actb | 9771 | -0.298 | -0.1975 | Yes |
| 83 | Mvd | 9923 | -0.315 | -0.1997 | Yes |
| 84 | Ldlrap1 | 9966 | -0.319 | -0.1922 | Yes |
| 85 | Gnai2 | 10068 | -0.332 | -0.1895 | Yes |
| 86 | Sorbs3 | 10081 | -0.332 | -0.1790 | Yes |
| 87 | Itga2 | 10098 | -0.334 | -0.1688 | Yes |
| 88 | Inppl1 | 10300 | -0.359 | -0.1738 | Yes |
| 89 | Rsu1 | 10389 | -0.373 | -0.1685 | Yes |
| 90 | Pik3cb | 10690 | -0.416 | -0.1801 | Yes |
| 91 | Mapk14 | 10704 | -0.420 | -0.1667 | Yes |
| 92 | Msn | 10802 | -0.434 | -0.1601 | Yes |
| 93 | Tjp1 | 10840 | -0.443 | -0.1479 | Yes |
| 94 | Cercam | 10950 | -0.471 | -0.1410 | Yes |
| 95 | Evl | 11042 | -0.490 | -0.1319 | Yes |
| 96 | Vcl | 11160 | -0.527 | -0.1238 | Yes |
| 97 | Tubg1 | 11345 | -0.597 | -0.1191 | Yes |
| 98 | Dlg1 | 11364 | -0.608 | -0.0995 | Yes |
| 99 | Actn1 | 11427 | -0.641 | -0.0826 | Yes |
| 100 | Syk | 11451 | -0.660 | -0.0617 | Yes |
| 101 | Sirpa | 11495 | -0.710 | -0.0408 | Yes |
| 102 | Adam9 | 11502 | -0.717 | -0.0164 | Yes |
| 103 | Pecam1 | 11517 | -0.739 | 0.0080 | Yes |