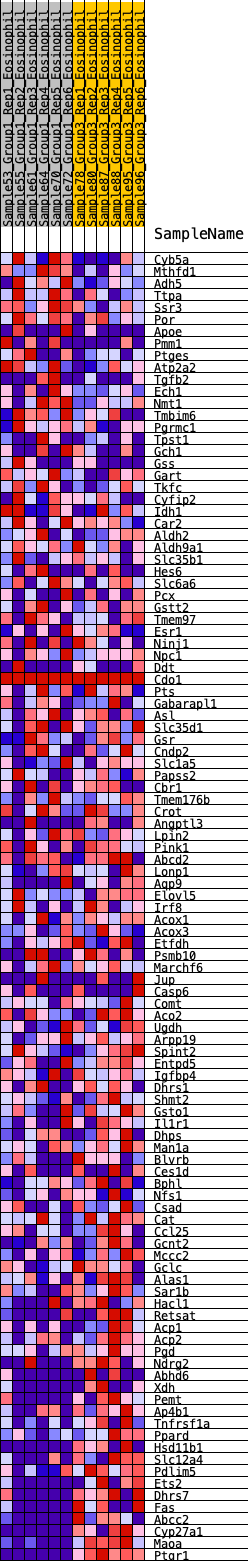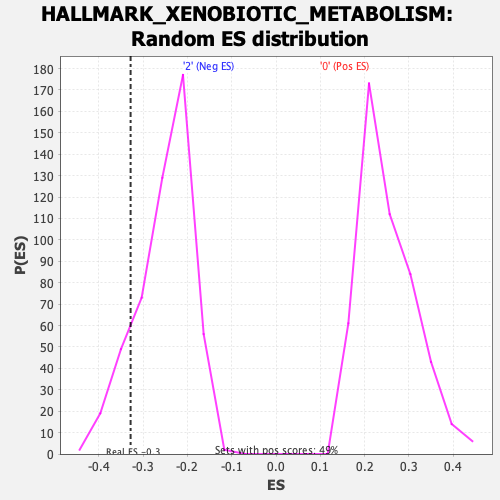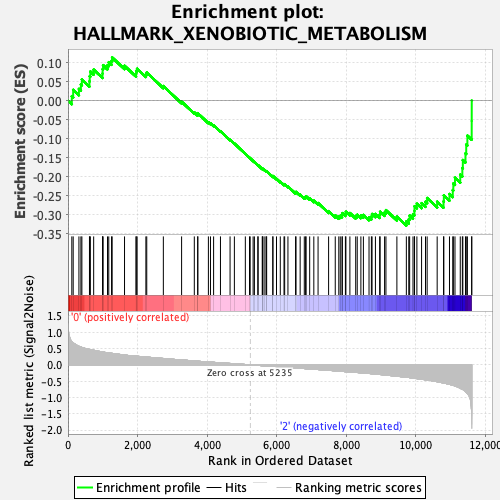
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.32830662 |
| Normalized Enrichment Score (NES) | -1.3014712 |
| Nominal p-value | 0.13017751 |
| FDR q-value | 0.32401693 |
| FWER p-Value | 0.76 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyb5a | 110 | 0.710 | 0.0115 | No |
| 2 | Mthfd1 | 149 | 0.675 | 0.0282 | No |
| 3 | Adh5 | 314 | 0.568 | 0.0307 | No |
| 4 | Ttpa | 371 | 0.545 | 0.0420 | No |
| 5 | Ssr3 | 399 | 0.533 | 0.0555 | No |
| 6 | Por | 617 | 0.472 | 0.0506 | No |
| 7 | Apoe | 624 | 0.470 | 0.0640 | No |
| 8 | Pmm1 | 642 | 0.467 | 0.0763 | No |
| 9 | Ptges | 738 | 0.445 | 0.0813 | No |
| 10 | Atp2a2 | 992 | 0.395 | 0.0710 | No |
| 11 | Tgfb2 | 995 | 0.394 | 0.0825 | No |
| 12 | Ech1 | 1009 | 0.391 | 0.0929 | No |
| 13 | Nmt1 | 1136 | 0.371 | 0.0930 | No |
| 14 | Tmbim6 | 1172 | 0.365 | 0.1008 | No |
| 15 | Pgrmc1 | 1252 | 0.354 | 0.1044 | No |
| 16 | Tpst1 | 1269 | 0.351 | 0.1134 | No |
| 17 | Gch1 | 1625 | 0.301 | 0.0914 | No |
| 18 | Gss | 1956 | 0.265 | 0.0706 | No |
| 19 | Gart | 1961 | 0.265 | 0.0781 | No |
| 20 | Tkfc | 1987 | 0.262 | 0.0837 | No |
| 21 | Cyfip2 | 2239 | 0.238 | 0.0689 | No |
| 22 | Idh1 | 2264 | 0.237 | 0.0738 | No |
| 23 | Car2 | 2741 | 0.195 | 0.0382 | No |
| 24 | Aldh2 | 3266 | 0.149 | -0.0029 | No |
| 25 | Aldh9a1 | 3631 | 0.118 | -0.0311 | No |
| 26 | Slc35b1 | 3729 | 0.110 | -0.0363 | No |
| 27 | Hes6 | 3732 | 0.110 | -0.0332 | No |
| 28 | Slc6a6 | 4036 | 0.088 | -0.0569 | No |
| 29 | Pcx | 4096 | 0.083 | -0.0596 | No |
| 30 | Gstt2 | 4186 | 0.076 | -0.0651 | No |
| 31 | Tmem97 | 4383 | 0.061 | -0.0803 | No |
| 32 | Esr1 | 4659 | 0.042 | -0.1030 | No |
| 33 | Ninj1 | 4783 | 0.034 | -0.1127 | No |
| 34 | Npc1 | 5098 | 0.010 | -0.1397 | No |
| 35 | Ddt | 5218 | 0.001 | -0.1500 | No |
| 36 | Cdo1 | 5237 | 0.000 | -0.1516 | No |
| 37 | Pts | 5322 | -0.006 | -0.1587 | No |
| 38 | Gabarapl1 | 5359 | -0.008 | -0.1616 | No |
| 39 | Asl | 5457 | -0.015 | -0.1695 | No |
| 40 | Slc35d1 | 5466 | -0.016 | -0.1698 | No |
| 41 | Gsr | 5587 | -0.026 | -0.1794 | No |
| 42 | Cndp2 | 5588 | -0.026 | -0.1787 | No |
| 43 | Slc1a5 | 5606 | -0.027 | -0.1794 | No |
| 44 | Papss2 | 5661 | -0.030 | -0.1832 | No |
| 45 | Cbr1 | 5700 | -0.033 | -0.1855 | No |
| 46 | Tmem176b | 5711 | -0.034 | -0.1854 | No |
| 47 | Crot | 5889 | -0.046 | -0.1994 | No |
| 48 | Angptl3 | 5890 | -0.046 | -0.1980 | No |
| 49 | Lpin2 | 5994 | -0.054 | -0.2054 | No |
| 50 | Pink1 | 6103 | -0.061 | -0.2130 | No |
| 51 | Abcd2 | 6210 | -0.070 | -0.2201 | No |
| 52 | Lonp1 | 6229 | -0.071 | -0.2196 | No |
| 53 | Aqp9 | 6323 | -0.078 | -0.2254 | No |
| 54 | Elovl5 | 6542 | -0.093 | -0.2416 | No |
| 55 | Irf8 | 6555 | -0.094 | -0.2398 | No |
| 56 | Acox1 | 6674 | -0.103 | -0.2470 | No |
| 57 | Acox3 | 6793 | -0.113 | -0.2539 | No |
| 58 | Etfdh | 6830 | -0.117 | -0.2536 | No |
| 59 | Psmb10 | 6845 | -0.118 | -0.2513 | No |
| 60 | Marchf6 | 6948 | -0.124 | -0.2565 | No |
| 61 | Jup | 7067 | -0.134 | -0.2628 | No |
| 62 | Casp6 | 7189 | -0.144 | -0.2691 | No |
| 63 | Comt | 7492 | -0.166 | -0.2904 | No |
| 64 | Aco2 | 7683 | -0.183 | -0.3015 | No |
| 65 | Ugdh | 7785 | -0.192 | -0.3046 | No |
| 66 | Arpp19 | 7827 | -0.195 | -0.3024 | No |
| 67 | Spint2 | 7878 | -0.200 | -0.3008 | No |
| 68 | Entpd5 | 7887 | -0.201 | -0.2956 | No |
| 69 | Igfbp4 | 7975 | -0.209 | -0.2970 | No |
| 70 | Dhrs1 | 7989 | -0.210 | -0.2919 | No |
| 71 | Shmt2 | 8104 | -0.218 | -0.2953 | No |
| 72 | Gsto1 | 8271 | -0.230 | -0.3029 | No |
| 73 | Il1r1 | 8315 | -0.235 | -0.2997 | No |
| 74 | Dhps | 8420 | -0.243 | -0.3016 | No |
| 75 | Man1a | 8492 | -0.250 | -0.3003 | No |
| 76 | Blvrb | 8654 | -0.265 | -0.3065 | No |
| 77 | Ces1d | 8722 | -0.270 | -0.3043 | No |
| 78 | Bphl | 8741 | -0.272 | -0.2978 | No |
| 79 | Nfs1 | 8838 | -0.283 | -0.2978 | No |
| 80 | Csad | 8960 | -0.295 | -0.2995 | No |
| 81 | Cat | 8973 | -0.296 | -0.2918 | No |
| 82 | Ccl25 | 9101 | -0.308 | -0.2937 | No |
| 83 | Gcnt2 | 9140 | -0.313 | -0.2877 | No |
| 84 | Mccc2 | 9455 | -0.347 | -0.3047 | No |
| 85 | Gclc | 9727 | -0.378 | -0.3171 | Yes |
| 86 | Alas1 | 9797 | -0.388 | -0.3116 | Yes |
| 87 | Sar1b | 9822 | -0.391 | -0.3021 | Yes |
| 88 | Hacl1 | 9918 | -0.407 | -0.2983 | Yes |
| 89 | Retsat | 9959 | -0.412 | -0.2896 | Yes |
| 90 | Acp1 | 9962 | -0.412 | -0.2775 | Yes |
| 91 | Acp2 | 10030 | -0.421 | -0.2709 | Yes |
| 92 | Pgd | 10165 | -0.443 | -0.2694 | Yes |
| 93 | Ndrg2 | 10278 | -0.457 | -0.2656 | Yes |
| 94 | Abhd6 | 10325 | -0.466 | -0.2558 | Yes |
| 95 | Xdh | 10613 | -0.518 | -0.2654 | Yes |
| 96 | Pemt | 10796 | -0.556 | -0.2648 | Yes |
| 97 | Ap4b1 | 10805 | -0.558 | -0.2489 | Yes |
| 98 | Tnfrsf1a | 10967 | -0.594 | -0.2453 | Yes |
| 99 | Ppard | 11062 | -0.623 | -0.2350 | Yes |
| 100 | Hsd11b1 | 11078 | -0.631 | -0.2176 | Yes |
| 101 | Slc12a4 | 11125 | -0.647 | -0.2025 | Yes |
| 102 | Pdlim5 | 11277 | -0.718 | -0.1943 | Yes |
| 103 | Ets2 | 11337 | -0.748 | -0.1773 | Yes |
| 104 | Dhrs7 | 11351 | -0.753 | -0.1561 | Yes |
| 105 | Fas | 11427 | -0.812 | -0.1386 | Yes |
| 106 | Abcc2 | 11449 | -0.844 | -0.1154 | Yes |
| 107 | Cyp27a1 | 11482 | -0.882 | -0.0920 | Yes |
| 108 | Maoa | 11606 | -1.656 | -0.0536 | Yes |
| 109 | Ptgr1 | 11607 | -1.816 | 0.0002 | Yes |