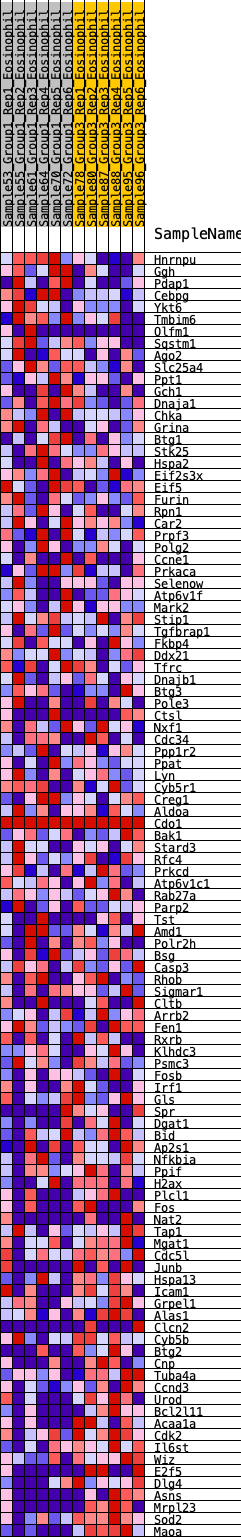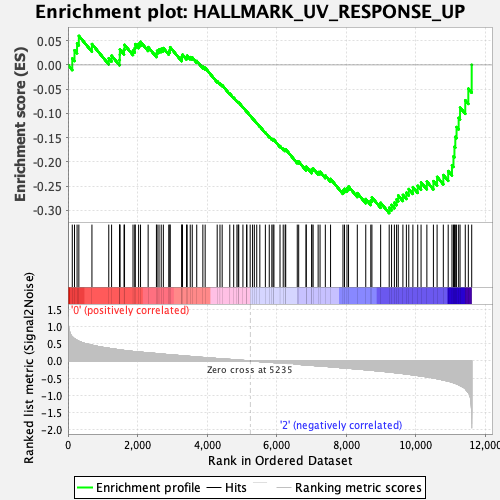
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.30555514 |
| Normalized Enrichment Score (NES) | -1.4154042 |
| Nominal p-value | 0.015355086 |
| FDR q-value | 0.48358628 |
| FWER p-Value | 0.558 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hnrnpu | 123 | 0.696 | 0.0134 | No |
| 2 | Ggh | 187 | 0.642 | 0.0302 | No |
| 3 | Pdap1 | 261 | 0.595 | 0.0446 | No |
| 4 | Cebpg | 312 | 0.569 | 0.0599 | No |
| 5 | Ykt6 | 688 | 0.456 | 0.0431 | No |
| 6 | Tmbim6 | 1172 | 0.365 | 0.0138 | No |
| 7 | Olfm1 | 1253 | 0.353 | 0.0191 | No |
| 8 | Sqstm1 | 1482 | 0.320 | 0.0104 | No |
| 9 | Ago2 | 1489 | 0.319 | 0.0210 | No |
| 10 | Slc25a4 | 1492 | 0.319 | 0.0318 | No |
| 11 | Ppt1 | 1611 | 0.303 | 0.0321 | No |
| 12 | Gch1 | 1625 | 0.301 | 0.0414 | No |
| 13 | Dnaja1 | 1869 | 0.273 | 0.0298 | No |
| 14 | Chka | 1920 | 0.268 | 0.0347 | No |
| 15 | Grina | 1933 | 0.267 | 0.0429 | No |
| 16 | Btg1 | 2030 | 0.258 | 0.0436 | No |
| 17 | Stk25 | 2085 | 0.253 | 0.0476 | No |
| 18 | Hspa2 | 2304 | 0.233 | 0.0368 | No |
| 19 | Eif2s3x | 2542 | 0.213 | 0.0236 | No |
| 20 | Eif5 | 2561 | 0.211 | 0.0293 | No |
| 21 | Furin | 2616 | 0.206 | 0.0318 | No |
| 22 | Rpn1 | 2680 | 0.200 | 0.0333 | No |
| 23 | Car2 | 2741 | 0.195 | 0.0348 | No |
| 24 | Prpf3 | 2897 | 0.180 | 0.0276 | No |
| 25 | Polg2 | 2929 | 0.177 | 0.0310 | No |
| 26 | Ccne1 | 2936 | 0.175 | 0.0366 | No |
| 27 | Prkaca | 3269 | 0.148 | 0.0128 | No |
| 28 | Selenow | 3274 | 0.147 | 0.0176 | No |
| 29 | Atp6v1f | 3292 | 0.146 | 0.0212 | No |
| 30 | Mark2 | 3408 | 0.137 | 0.0160 | No |
| 31 | Stip1 | 3425 | 0.136 | 0.0193 | No |
| 32 | Tgfbrap1 | 3514 | 0.129 | 0.0161 | No |
| 33 | Fkbp4 | 3572 | 0.123 | 0.0154 | No |
| 34 | Ddx21 | 3701 | 0.112 | 0.0082 | No |
| 35 | Tfrc | 3877 | 0.100 | -0.0036 | No |
| 36 | Dnajb1 | 3943 | 0.095 | -0.0059 | No |
| 37 | Btg3 | 4289 | 0.068 | -0.0336 | No |
| 38 | Pole3 | 4369 | 0.061 | -0.0383 | No |
| 39 | Ctsl | 4432 | 0.057 | -0.0417 | No |
| 40 | Nxf1 | 4651 | 0.043 | -0.0591 | No |
| 41 | Cdc34 | 4763 | 0.036 | -0.0676 | No |
| 42 | Ppp1r2 | 4857 | 0.028 | -0.0747 | No |
| 43 | Ppat | 4893 | 0.025 | -0.0768 | No |
| 44 | Lyn | 4909 | 0.024 | -0.0773 | No |
| 45 | Cyb5r1 | 5029 | 0.015 | -0.0872 | No |
| 46 | Creg1 | 5134 | 0.007 | -0.0960 | No |
| 47 | Aldoa | 5143 | 0.006 | -0.0964 | No |
| 48 | Cdo1 | 5237 | 0.000 | -0.1045 | No |
| 49 | Bak1 | 5306 | -0.005 | -0.1103 | No |
| 50 | Stard3 | 5357 | -0.008 | -0.1143 | No |
| 51 | Rfc4 | 5426 | -0.013 | -0.1198 | No |
| 52 | Prkcd | 5515 | -0.020 | -0.1267 | No |
| 53 | Atp6v1c1 | 5674 | -0.030 | -0.1394 | No |
| 54 | Rab27a | 5786 | -0.039 | -0.1477 | No |
| 55 | Parp2 | 5860 | -0.044 | -0.1526 | No |
| 56 | Tst | 5895 | -0.047 | -0.1539 | No |
| 57 | Amd1 | 5920 | -0.049 | -0.1543 | No |
| 58 | Polr2h | 6095 | -0.061 | -0.1673 | No |
| 59 | Bsg | 6187 | -0.067 | -0.1729 | No |
| 60 | Casp3 | 6242 | -0.072 | -0.1751 | No |
| 61 | Rhob | 6259 | -0.073 | -0.1739 | No |
| 62 | Sigmar1 | 6591 | -0.098 | -0.1993 | No |
| 63 | Cltb | 6632 | -0.100 | -0.1993 | No |
| 64 | Arrb2 | 6844 | -0.118 | -0.2136 | No |
| 65 | Fen1 | 6848 | -0.118 | -0.2097 | No |
| 66 | Rxrb | 7005 | -0.128 | -0.2189 | No |
| 67 | Klhdc3 | 7010 | -0.128 | -0.2148 | No |
| 68 | Psmc3 | 7047 | -0.132 | -0.2133 | No |
| 69 | Fosb | 7191 | -0.144 | -0.2208 | No |
| 70 | Irf1 | 7246 | -0.148 | -0.2203 | No |
| 71 | Gls | 7398 | -0.159 | -0.2280 | No |
| 72 | Spr | 7548 | -0.171 | -0.2350 | No |
| 73 | Dgat1 | 7905 | -0.202 | -0.2589 | No |
| 74 | Bid | 7949 | -0.206 | -0.2555 | No |
| 75 | Ap2s1 | 8020 | -0.211 | -0.2543 | No |
| 76 | Nfkbia | 8065 | -0.215 | -0.2506 | No |
| 77 | Ppif | 8317 | -0.235 | -0.2643 | No |
| 78 | H2ax | 8560 | -0.257 | -0.2764 | No |
| 79 | Plcl1 | 8705 | -0.269 | -0.2796 | No |
| 80 | Fos | 8737 | -0.272 | -0.2729 | No |
| 81 | Nat2 | 8988 | -0.298 | -0.2843 | No |
| 82 | Tap1 | 9234 | -0.324 | -0.2943 | Yes |
| 83 | Mgat1 | 9302 | -0.332 | -0.2886 | Yes |
| 84 | Cdc5l | 9383 | -0.339 | -0.2838 | Yes |
| 85 | Junb | 9446 | -0.347 | -0.2772 | Yes |
| 86 | Hspa13 | 9494 | -0.352 | -0.2691 | Yes |
| 87 | Icam1 | 9628 | -0.367 | -0.2679 | Yes |
| 88 | Grpel1 | 9728 | -0.378 | -0.2634 | Yes |
| 89 | Alas1 | 9797 | -0.388 | -0.2558 | Yes |
| 90 | Clcn2 | 9915 | -0.407 | -0.2519 | Yes |
| 91 | Cyb5b | 10054 | -0.424 | -0.2492 | Yes |
| 92 | Btg2 | 10150 | -0.441 | -0.2421 | Yes |
| 93 | Cnp | 10317 | -0.465 | -0.2404 | Yes |
| 94 | Tuba4a | 10505 | -0.494 | -0.2396 | Yes |
| 95 | Ccnd3 | 10611 | -0.517 | -0.2308 | Yes |
| 96 | Urod | 10790 | -0.555 | -0.2270 | Yes |
| 97 | Bcl2l11 | 10930 | -0.585 | -0.2188 | Yes |
| 98 | Acaa1a | 11039 | -0.618 | -0.2067 | Yes |
| 99 | Cdk2 | 11082 | -0.632 | -0.1884 | Yes |
| 100 | Il6st | 11110 | -0.641 | -0.1685 | Yes |
| 101 | Wiz | 11135 | -0.652 | -0.1480 | Yes |
| 102 | E2f5 | 11171 | -0.665 | -0.1280 | Yes |
| 103 | Dlg4 | 11231 | -0.691 | -0.1091 | Yes |
| 104 | Asns | 11271 | -0.712 | -0.0878 | Yes |
| 105 | Mrpl23 | 11421 | -0.803 | -0.0729 | Yes |
| 106 | Sod2 | 11509 | -0.912 | -0.0488 | Yes |
| 107 | Maoa | 11606 | -1.656 | 0.0003 | Yes |