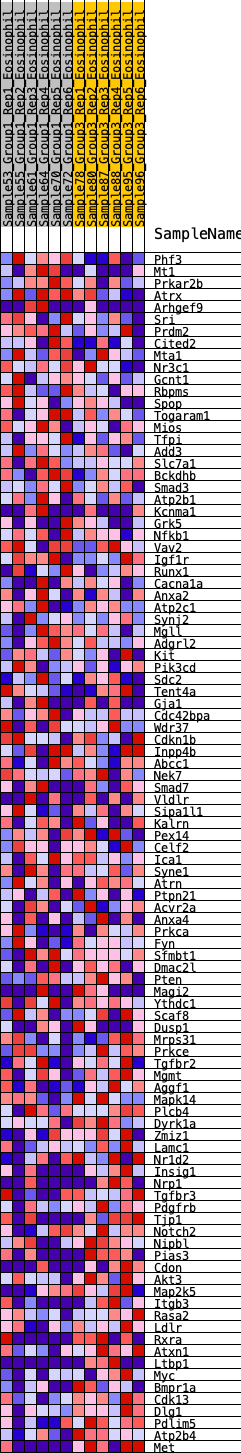
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
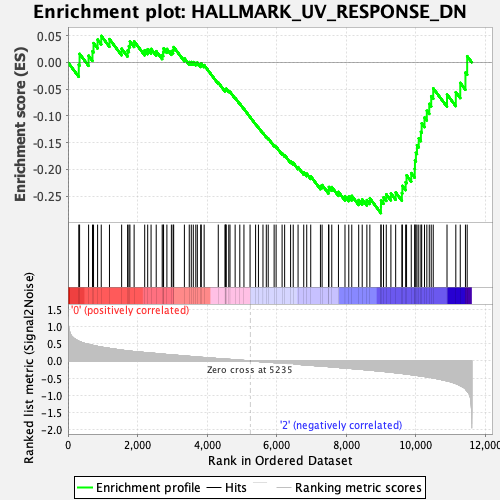
| GeneSet | HALLMARK_UV_RESPONSE_DN |
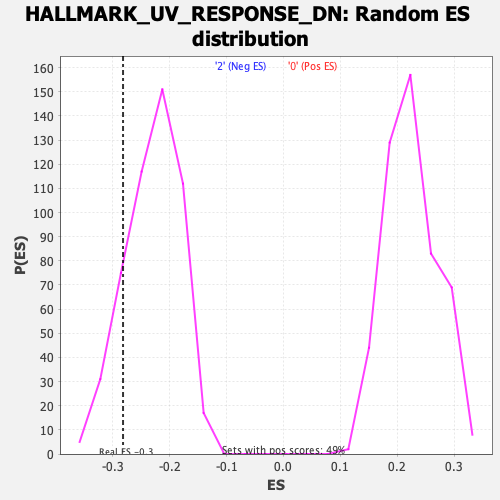
| Enrichment Score (ES) | -0.2818725 |
| Normalized Enrichment Score (NES) | -1.2307631 |
| Nominal p-value | 0.15354331 |
| FDR q-value | 0.38854408 |
| FWER p-Value | 0.864 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Phf3 | 310 | 0.569 | -0.0043 | No |
| 2 | Mt1 | 333 | 0.560 | 0.0160 | No |
| 3 | Prkar2b | 592 | 0.479 | 0.0126 | No |
| 4 | Atrx | 704 | 0.452 | 0.0209 | No |
| 5 | Arhgef9 | 734 | 0.446 | 0.0361 | No |
| 6 | Sri | 851 | 0.420 | 0.0427 | No |
| 7 | Prdm2 | 956 | 0.402 | 0.0496 | No |
| 8 | Cited2 | 1192 | 0.362 | 0.0436 | No |
| 9 | Mta1 | 1542 | 0.312 | 0.0256 | No |
| 10 | Nr3c1 | 1713 | 0.288 | 0.0223 | No |
| 11 | Gcnt1 | 1751 | 0.285 | 0.0304 | No |
| 12 | Rbpms | 1780 | 0.282 | 0.0392 | No |
| 13 | Spop | 1901 | 0.270 | 0.0395 | No |
| 14 | Togaram1 | 2205 | 0.242 | 0.0227 | No |
| 15 | Mios | 2291 | 0.234 | 0.0246 | No |
| 16 | Tfpi | 2388 | 0.225 | 0.0252 | No |
| 17 | Add3 | 2536 | 0.214 | 0.0210 | No |
| 18 | Slc7a1 | 2708 | 0.197 | 0.0139 | No |
| 19 | Bckdhb | 2737 | 0.195 | 0.0192 | No |
| 20 | Smad3 | 2747 | 0.194 | 0.0262 | No |
| 21 | Atp2b1 | 2843 | 0.184 | 0.0252 | No |
| 22 | Kcnma1 | 2972 | 0.173 | 0.0209 | No |
| 23 | Grk5 | 3021 | 0.169 | 0.0235 | No |
| 24 | Nfkb1 | 3038 | 0.167 | 0.0287 | No |
| 25 | Vav2 | 3346 | 0.142 | 0.0077 | No |
| 26 | Igf1r | 3486 | 0.130 | 0.0008 | No |
| 27 | Runx1 | 3544 | 0.126 | 0.0008 | No |
| 28 | Cacna1a | 3602 | 0.120 | 0.0006 | No |
| 29 | Anxa2 | 3674 | 0.115 | -0.0010 | No |
| 30 | Atp2c1 | 3718 | 0.111 | -0.0003 | No |
| 31 | Synj2 | 3815 | 0.104 | -0.0046 | No |
| 32 | Mgll | 3831 | 0.103 | -0.0018 | No |
| 33 | Adgrl2 | 3914 | 0.097 | -0.0051 | No |
| 34 | Kit | 4320 | 0.065 | -0.0377 | No |
| 35 | Pik3cd | 4514 | 0.052 | -0.0524 | No |
| 36 | Sdc2 | 4525 | 0.051 | -0.0512 | No |
| 37 | Tent4a | 4538 | 0.050 | -0.0503 | No |
| 38 | Gja1 | 4547 | 0.049 | -0.0490 | No |
| 39 | Cdc42bpa | 4622 | 0.044 | -0.0537 | No |
| 40 | Wdr37 | 4657 | 0.043 | -0.0550 | No |
| 41 | Cdkn1b | 4805 | 0.032 | -0.0665 | No |
| 42 | Inpp4b | 4939 | 0.021 | -0.0772 | No |
| 43 | Abcc1 | 5058 | 0.013 | -0.0869 | No |
| 44 | Nek7 | 5234 | 0.000 | -0.1021 | No |
| 45 | Smad7 | 5393 | -0.011 | -0.1154 | No |
| 46 | Vldlr | 5475 | -0.017 | -0.1218 | No |
| 47 | Sipa1l1 | 5605 | -0.026 | -0.1319 | No |
| 48 | Kalrn | 5701 | -0.033 | -0.1389 | No |
| 49 | Pex14 | 5756 | -0.037 | -0.1421 | No |
| 50 | Celf2 | 5926 | -0.049 | -0.1549 | No |
| 51 | Ica1 | 5983 | -0.053 | -0.1576 | No |
| 52 | Syne1 | 6155 | -0.065 | -0.1699 | No |
| 53 | Atrn | 6228 | -0.071 | -0.1733 | No |
| 54 | Ptpn21 | 6399 | -0.083 | -0.1848 | No |
| 55 | Acvr2a | 6475 | -0.090 | -0.1878 | No |
| 56 | Anxa4 | 6614 | -0.099 | -0.1958 | No |
| 57 | Prkca | 6777 | -0.112 | -0.2054 | No |
| 58 | Fyn | 6863 | -0.120 | -0.2081 | No |
| 59 | Sfmbt1 | 6978 | -0.127 | -0.2130 | No |
| 60 | Dmac2l | 7257 | -0.149 | -0.2312 | No |
| 61 | Pten | 7305 | -0.152 | -0.2293 | No |
| 62 | Magi2 | 7490 | -0.165 | -0.2387 | No |
| 63 | Ythdc1 | 7500 | -0.166 | -0.2329 | No |
| 64 | Scaf8 | 7583 | -0.174 | -0.2331 | No |
| 65 | Dusp1 | 7775 | -0.191 | -0.2421 | No |
| 66 | Mrps31 | 7963 | -0.207 | -0.2501 | No |
| 67 | Prkce | 8072 | -0.216 | -0.2509 | No |
| 68 | Tgfbr2 | 8158 | -0.222 | -0.2495 | No |
| 69 | Mgmt | 8357 | -0.238 | -0.2573 | No |
| 70 | Aggf1 | 8458 | -0.247 | -0.2562 | No |
| 71 | Mapk14 | 8592 | -0.259 | -0.2574 | No |
| 72 | Plcb4 | 8678 | -0.266 | -0.2542 | No |
| 73 | Dyrk1a | 8997 | -0.298 | -0.2700 | Yes |
| 74 | Zmiz1 | 8999 | -0.298 | -0.2583 | Yes |
| 75 | Lamc1 | 9071 | -0.305 | -0.2523 | Yes |
| 76 | Nr1d2 | 9150 | -0.314 | -0.2466 | Yes |
| 77 | Insig1 | 9283 | -0.330 | -0.2450 | Yes |
| 78 | Nrp1 | 9420 | -0.344 | -0.2432 | Yes |
| 79 | Tgfbr3 | 9599 | -0.364 | -0.2442 | Yes |
| 80 | Pdgfrb | 9613 | -0.365 | -0.2308 | Yes |
| 81 | Tjp1 | 9707 | -0.376 | -0.2240 | Yes |
| 82 | Notch2 | 9733 | -0.378 | -0.2111 | Yes |
| 83 | Nipbl | 9868 | -0.399 | -0.2070 | Yes |
| 84 | Pias3 | 9968 | -0.413 | -0.1991 | Yes |
| 85 | Cdon | 9975 | -0.415 | -0.1832 | Yes |
| 86 | Akt3 | 10002 | -0.418 | -0.1689 | Yes |
| 87 | Map2k5 | 10033 | -0.422 | -0.1547 | Yes |
| 88 | Itgb3 | 10084 | -0.430 | -0.1420 | Yes |
| 89 | Rasa2 | 10146 | -0.440 | -0.1299 | Yes |
| 90 | Ldlr | 10168 | -0.443 | -0.1141 | Yes |
| 91 | Rxra | 10249 | -0.453 | -0.1030 | Yes |
| 92 | Atxn1 | 10316 | -0.465 | -0.0903 | Yes |
| 93 | Ltbp1 | 10386 | -0.474 | -0.0775 | Yes |
| 94 | Myc | 10442 | -0.482 | -0.0632 | Yes |
| 95 | Bmpr1a | 10500 | -0.493 | -0.0485 | Yes |
| 96 | Cdk13 | 10896 | -0.578 | -0.0599 | Yes |
| 97 | Dlg1 | 11149 | -0.656 | -0.0557 | Yes |
| 98 | Pdlim5 | 11277 | -0.718 | -0.0383 | Yes |
| 99 | Atp2b4 | 11426 | -0.811 | -0.0189 | Yes |
| 100 | Met | 11476 | -0.876 | 0.0116 | Yes |