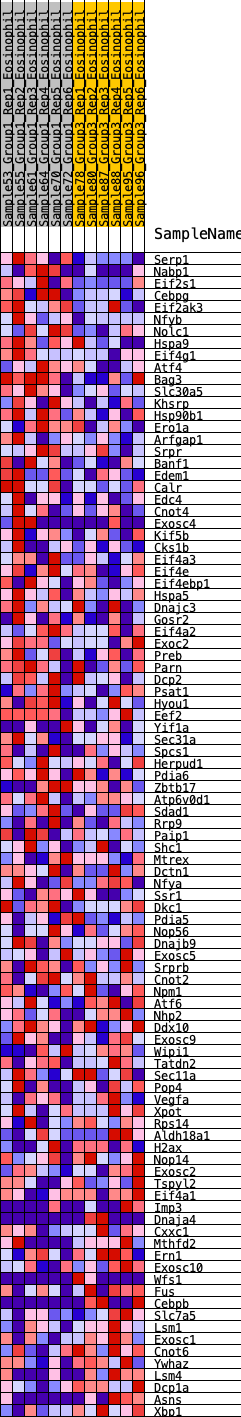
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
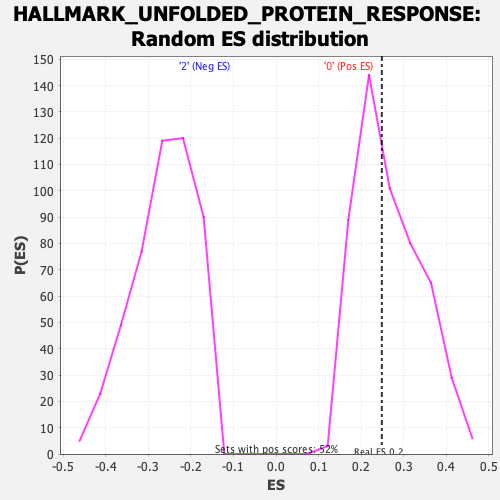
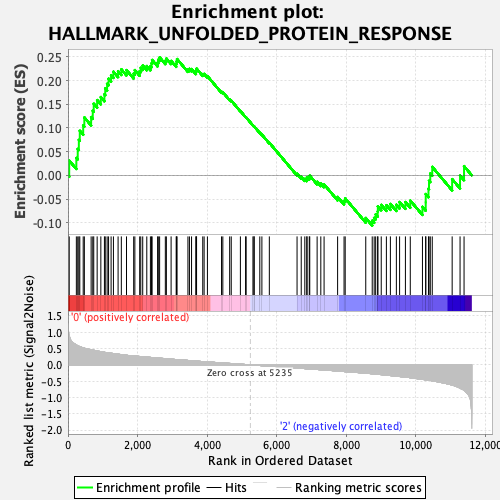
| Enrichment Score (ES) | 0.24877287 |
| Normalized Enrichment Score (NES) | 0.9365809 |
| Nominal p-value | 0.516441 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Serp1 | 35 | 0.891 | 0.0311 | Yes |
| 2 | Nabp1 | 240 | 0.609 | 0.0368 | Yes |
| 3 | Eif2s1 | 280 | 0.585 | 0.0558 | Yes |
| 4 | Cebpg | 312 | 0.569 | 0.0749 | Yes |
| 5 | Eif2ak3 | 339 | 0.558 | 0.0940 | Yes |
| 6 | Nfyb | 437 | 0.518 | 0.1054 | Yes |
| 7 | Nolc1 | 470 | 0.508 | 0.1221 | Yes |
| 8 | Hspa9 | 665 | 0.462 | 0.1230 | Yes |
| 9 | Eif4g1 | 712 | 0.451 | 0.1363 | Yes |
| 10 | Atf4 | 740 | 0.445 | 0.1510 | Yes |
| 11 | Bag3 | 838 | 0.422 | 0.1588 | Yes |
| 12 | Slc30a5 | 944 | 0.405 | 0.1652 | Yes |
| 13 | Khsrp | 1047 | 0.384 | 0.1710 | Yes |
| 14 | Hsp90b1 | 1071 | 0.379 | 0.1836 | Yes |
| 15 | Ero1a | 1125 | 0.373 | 0.1933 | Yes |
| 16 | Arfgap1 | 1166 | 0.366 | 0.2038 | Yes |
| 17 | Srpr | 1239 | 0.355 | 0.2112 | Yes |
| 18 | Banf1 | 1306 | 0.345 | 0.2187 | Yes |
| 19 | Edem1 | 1440 | 0.326 | 0.2196 | Yes |
| 20 | Calr | 1533 | 0.313 | 0.2236 | Yes |
| 21 | Edc4 | 1682 | 0.293 | 0.2220 | Yes |
| 22 | Cnot4 | 1888 | 0.272 | 0.2146 | Yes |
| 23 | Exosc4 | 1922 | 0.268 | 0.2220 | Yes |
| 24 | Kif5b | 2061 | 0.255 | 0.2198 | Yes |
| 25 | Cks1b | 2092 | 0.253 | 0.2269 | Yes |
| 26 | Eif4a3 | 2148 | 0.247 | 0.2316 | Yes |
| 27 | Eif4e | 2265 | 0.237 | 0.2306 | Yes |
| 28 | Eif4ebp1 | 2369 | 0.227 | 0.2303 | Yes |
| 29 | Hspa5 | 2400 | 0.224 | 0.2363 | Yes |
| 30 | Dnajc3 | 2418 | 0.223 | 0.2434 | Yes |
| 31 | Gosr2 | 2576 | 0.210 | 0.2378 | Yes |
| 32 | Eif4a2 | 2596 | 0.208 | 0.2441 | Yes |
| 33 | Exoc2 | 2634 | 0.205 | 0.2488 | Yes |
| 34 | Preb | 2798 | 0.189 | 0.2419 | No |
| 35 | Parn | 2828 | 0.185 | 0.2464 | No |
| 36 | Dcp2 | 2964 | 0.173 | 0.2414 | No |
| 37 | Psat1 | 3110 | 0.162 | 0.2350 | No |
| 38 | Hyou1 | 3120 | 0.161 | 0.2404 | No |
| 39 | Eef2 | 3135 | 0.160 | 0.2453 | No |
| 40 | Yif1a | 3448 | 0.134 | 0.2233 | No |
| 41 | Sec31a | 3491 | 0.130 | 0.2247 | No |
| 42 | Spcs1 | 3552 | 0.125 | 0.2243 | No |
| 43 | Herpud1 | 3677 | 0.115 | 0.2179 | No |
| 44 | Pdia6 | 3684 | 0.114 | 0.2217 | No |
| 45 | Zbtb17 | 3696 | 0.113 | 0.2251 | No |
| 46 | Atp6v0d1 | 3874 | 0.100 | 0.2136 | No |
| 47 | Sdad1 | 3913 | 0.097 | 0.2140 | No |
| 48 | Rrp9 | 4008 | 0.090 | 0.2093 | No |
| 49 | Paip1 | 4414 | 0.059 | 0.1763 | No |
| 50 | Shc1 | 4451 | 0.056 | 0.1754 | No |
| 51 | Mtrex | 4648 | 0.043 | 0.1600 | No |
| 52 | Dctn1 | 4693 | 0.040 | 0.1577 | No |
| 53 | Nfya | 4956 | 0.019 | 0.1357 | No |
| 54 | Ssr1 | 5105 | 0.009 | 0.1232 | No |
| 55 | Dkc1 | 5121 | 0.008 | 0.1222 | No |
| 56 | Pdia5 | 5317 | -0.006 | 0.1055 | No |
| 57 | Nop56 | 5348 | -0.008 | 0.1032 | No |
| 58 | Dnajb9 | 5351 | -0.008 | 0.1033 | No |
| 59 | Exosc5 | 5514 | -0.020 | 0.0900 | No |
| 60 | Srprb | 5574 | -0.025 | 0.0858 | No |
| 61 | Cnot2 | 5788 | -0.039 | 0.0688 | No |
| 62 | Npm1 | 6585 | -0.097 | 0.0034 | No |
| 63 | Atf6 | 6705 | -0.106 | -0.0029 | No |
| 64 | Nhp2 | 6808 | -0.115 | -0.0074 | No |
| 65 | Ddx10 | 6865 | -0.120 | -0.0076 | No |
| 66 | Exosc9 | 6872 | -0.120 | -0.0036 | No |
| 67 | Wipi1 | 6936 | -0.123 | -0.0043 | No |
| 68 | Tatdn2 | 6950 | -0.125 | -0.0007 | No |
| 69 | Sec11a | 7162 | -0.141 | -0.0136 | No |
| 70 | Pop4 | 7268 | -0.149 | -0.0170 | No |
| 71 | Vegfa | 7362 | -0.156 | -0.0191 | No |
| 72 | Xpot | 7751 | -0.188 | -0.0456 | No |
| 73 | Rps14 | 7936 | -0.205 | -0.0537 | No |
| 74 | Aldh18a1 | 7969 | -0.208 | -0.0485 | No |
| 75 | H2ax | 8560 | -0.257 | -0.0899 | No |
| 76 | Nop14 | 8746 | -0.273 | -0.0955 | No |
| 77 | Exosc2 | 8802 | -0.279 | -0.0896 | No |
| 78 | Tspyl2 | 8845 | -0.284 | -0.0824 | No |
| 79 | Eif4a1 | 8903 | -0.290 | -0.0762 | No |
| 80 | Imp3 | 8908 | -0.291 | -0.0654 | No |
| 81 | Dnaja4 | 9005 | -0.299 | -0.0623 | No |
| 82 | Cxxc1 | 9153 | -0.315 | -0.0630 | No |
| 83 | Mthfd2 | 9268 | -0.328 | -0.0604 | No |
| 84 | Ern1 | 9440 | -0.346 | -0.0619 | No |
| 85 | Exosc10 | 9533 | -0.357 | -0.0563 | No |
| 86 | Wfs1 | 9700 | -0.375 | -0.0563 | No |
| 87 | Fus | 9840 | -0.395 | -0.0532 | No |
| 88 | Cebpb | 10190 | -0.447 | -0.0664 | No |
| 89 | Slc7a5 | 10284 | -0.457 | -0.0570 | No |
| 90 | Lsm1 | 10285 | -0.458 | -0.0394 | No |
| 91 | Exosc1 | 10363 | -0.470 | -0.0281 | No |
| 92 | Cnot6 | 10379 | -0.472 | -0.0113 | No |
| 93 | Ywhaz | 10415 | -0.479 | 0.0040 | No |
| 94 | Lsm4 | 10472 | -0.488 | 0.0179 | No |
| 95 | Dcp1a | 11044 | -0.618 | -0.0080 | No |
| 96 | Asns | 11271 | -0.712 | -0.0004 | No |
| 97 | Xbp1 | 11386 | -0.774 | 0.0194 | No |