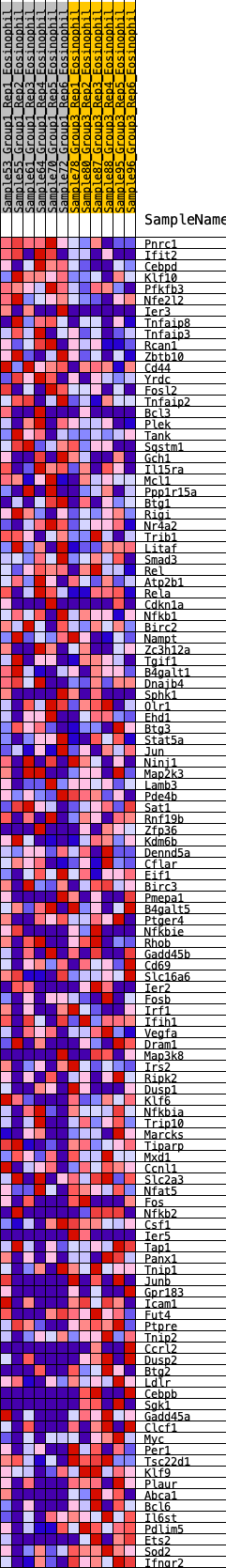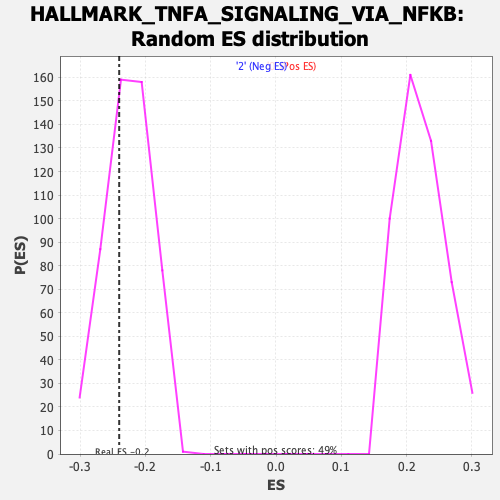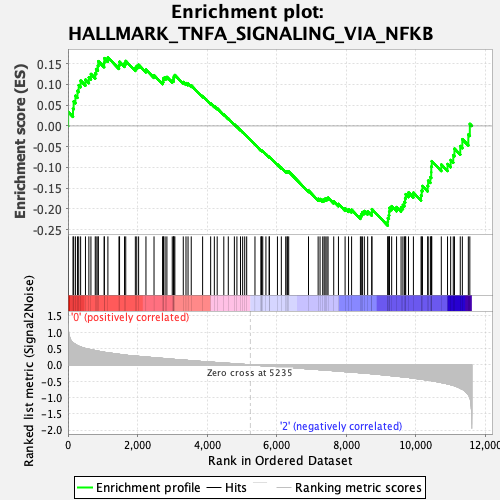
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | -0.24009469 |
| Normalized Enrichment Score (NES) | -1.0651323 |
| Nominal p-value | 0.30769232 |
| FDR q-value | 0.7088886 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pnrc1 | 4 | 1.194 | 0.0341 | No |
| 2 | Ifit2 | 144 | 0.678 | 0.0415 | No |
| 3 | Cebpd | 163 | 0.662 | 0.0590 | No |
| 4 | Klf10 | 213 | 0.625 | 0.0728 | No |
| 5 | Pfkfb3 | 272 | 0.588 | 0.0847 | No |
| 6 | Nfe2l2 | 308 | 0.570 | 0.0981 | No |
| 7 | Ier3 | 363 | 0.550 | 0.1092 | No |
| 8 | Tnfaip8 | 502 | 0.499 | 0.1116 | No |
| 9 | Tnfaip3 | 600 | 0.476 | 0.1169 | No |
| 10 | Rcan1 | 659 | 0.463 | 0.1252 | No |
| 11 | Zbtb10 | 783 | 0.435 | 0.1271 | No |
| 12 | Cd44 | 809 | 0.429 | 0.1373 | No |
| 13 | Yrdc | 859 | 0.419 | 0.1451 | No |
| 14 | Fosl2 | 872 | 0.417 | 0.1560 | No |
| 15 | Tnfaip2 | 1039 | 0.385 | 0.1527 | No |
| 16 | Bcl3 | 1046 | 0.384 | 0.1633 | No |
| 17 | Plek | 1147 | 0.369 | 0.1652 | No |
| 18 | Tank | 1466 | 0.321 | 0.1468 | No |
| 19 | Sqstm1 | 1482 | 0.320 | 0.1547 | No |
| 20 | Gch1 | 1625 | 0.301 | 0.1510 | No |
| 21 | Il15ra | 1660 | 0.296 | 0.1566 | No |
| 22 | Mcl1 | 1936 | 0.267 | 0.1404 | No |
| 23 | Ppp1r15a | 1976 | 0.263 | 0.1446 | No |
| 24 | Btg1 | 2030 | 0.258 | 0.1474 | No |
| 25 | Rigi | 2241 | 0.238 | 0.1360 | No |
| 26 | Nr4a2 | 2473 | 0.219 | 0.1222 | No |
| 27 | Trib1 | 2719 | 0.196 | 0.1065 | No |
| 28 | Litaf | 2738 | 0.195 | 0.1106 | No |
| 29 | Smad3 | 2747 | 0.194 | 0.1155 | No |
| 30 | Rel | 2791 | 0.190 | 0.1172 | No |
| 31 | Atp2b1 | 2843 | 0.184 | 0.1181 | No |
| 32 | Rela | 2997 | 0.171 | 0.1097 | No |
| 33 | Cdkn1a | 3035 | 0.167 | 0.1113 | No |
| 34 | Nfkb1 | 3038 | 0.167 | 0.1159 | No |
| 35 | Birc2 | 3046 | 0.167 | 0.1201 | No |
| 36 | Nampt | 3072 | 0.165 | 0.1227 | No |
| 37 | Zc3h12a | 3314 | 0.144 | 0.1059 | No |
| 38 | Tgif1 | 3392 | 0.139 | 0.1032 | No |
| 39 | B4galt1 | 3451 | 0.133 | 0.1020 | No |
| 40 | Dnajb4 | 3543 | 0.126 | 0.0977 | No |
| 41 | Sphk1 | 3870 | 0.100 | 0.0722 | No |
| 42 | Olr1 | 4101 | 0.083 | 0.0546 | No |
| 43 | Ehd1 | 4207 | 0.075 | 0.0476 | No |
| 44 | Btg3 | 4289 | 0.068 | 0.0425 | No |
| 45 | Stat5a | 4479 | 0.055 | 0.0276 | No |
| 46 | Jun | 4608 | 0.045 | 0.0178 | No |
| 47 | Ninj1 | 4783 | 0.034 | 0.0037 | No |
| 48 | Map2k3 | 4856 | 0.028 | -0.0018 | No |
| 49 | Lamb3 | 4959 | 0.019 | -0.0101 | No |
| 50 | Pde4b | 5021 | 0.015 | -0.0150 | No |
| 51 | Sat1 | 5080 | 0.012 | -0.0197 | No |
| 52 | Rnf19b | 5137 | 0.007 | -0.0244 | No |
| 53 | Zfp36 | 5375 | -0.009 | -0.0447 | No |
| 54 | Kdm6b | 5545 | -0.023 | -0.0588 | No |
| 55 | Dennd5a | 5561 | -0.023 | -0.0594 | No |
| 56 | Cflar | 5572 | -0.025 | -0.0596 | No |
| 57 | Eif1 | 5599 | -0.026 | -0.0611 | No |
| 58 | Birc3 | 5690 | -0.032 | -0.0680 | No |
| 59 | Pmepa1 | 5782 | -0.039 | -0.0748 | No |
| 60 | B4galt5 | 5791 | -0.039 | -0.0744 | No |
| 61 | Ptger4 | 6021 | -0.056 | -0.0927 | No |
| 62 | Nfkbie | 6133 | -0.063 | -0.1005 | No |
| 63 | Rhob | 6259 | -0.073 | -0.1093 | No |
| 64 | Gadd45b | 6295 | -0.076 | -0.1102 | No |
| 65 | Cd69 | 6314 | -0.077 | -0.1095 | No |
| 66 | Slc16a6 | 6345 | -0.080 | -0.1098 | No |
| 67 | Ier2 | 6914 | -0.122 | -0.1557 | No |
| 68 | Fosb | 7191 | -0.144 | -0.1756 | No |
| 69 | Irf1 | 7246 | -0.148 | -0.1760 | No |
| 70 | Ifih1 | 7323 | -0.153 | -0.1782 | No |
| 71 | Vegfa | 7362 | -0.156 | -0.1770 | No |
| 72 | Dram1 | 7393 | -0.158 | -0.1751 | No |
| 73 | Map3k8 | 7442 | -0.161 | -0.1746 | No |
| 74 | Irs2 | 7476 | -0.164 | -0.1727 | No |
| 75 | Ripk2 | 7639 | -0.179 | -0.1817 | No |
| 76 | Dusp1 | 7775 | -0.191 | -0.1879 | No |
| 77 | Klf6 | 7966 | -0.208 | -0.1985 | No |
| 78 | Nfkbia | 8065 | -0.215 | -0.2008 | No |
| 79 | Trip10 | 8153 | -0.221 | -0.2020 | No |
| 80 | Marcks | 8406 | -0.242 | -0.2169 | No |
| 81 | Tiparp | 8432 | -0.244 | -0.2121 | No |
| 82 | Mxd1 | 8464 | -0.248 | -0.2076 | No |
| 83 | Ccnl1 | 8519 | -0.253 | -0.2050 | No |
| 84 | Slc2a3 | 8615 | -0.261 | -0.2057 | No |
| 85 | Nfat5 | 8733 | -0.272 | -0.2081 | No |
| 86 | Fos | 8737 | -0.272 | -0.2005 | No |
| 87 | Nfkb2 | 9193 | -0.319 | -0.2309 | Yes |
| 88 | Csf1 | 9197 | -0.320 | -0.2219 | Yes |
| 89 | Ier5 | 9225 | -0.323 | -0.2149 | Yes |
| 90 | Tap1 | 9234 | -0.324 | -0.2063 | Yes |
| 91 | Panx1 | 9241 | -0.325 | -0.1974 | Yes |
| 92 | Tnip1 | 9305 | -0.332 | -0.1933 | Yes |
| 93 | Junb | 9446 | -0.347 | -0.1955 | Yes |
| 94 | Gpr183 | 9575 | -0.362 | -0.1962 | Yes |
| 95 | Icam1 | 9628 | -0.367 | -0.1901 | Yes |
| 96 | Fut4 | 9671 | -0.373 | -0.1831 | Yes |
| 97 | Ptpre | 9693 | -0.375 | -0.1741 | Yes |
| 98 | Tnip2 | 9708 | -0.376 | -0.1645 | Yes |
| 99 | Ccrl2 | 9789 | -0.387 | -0.1603 | Yes |
| 100 | Dusp2 | 9930 | -0.408 | -0.1607 | Yes |
| 101 | Btg2 | 10150 | -0.441 | -0.1670 | Yes |
| 102 | Ldlr | 10168 | -0.443 | -0.1557 | Yes |
| 103 | Cebpb | 10190 | -0.447 | -0.1447 | Yes |
| 104 | Sgk1 | 10340 | -0.467 | -0.1442 | Yes |
| 105 | Gadd45a | 10354 | -0.469 | -0.1318 | Yes |
| 106 | Clcf1 | 10417 | -0.479 | -0.1234 | Yes |
| 107 | Myc | 10442 | -0.482 | -0.1116 | Yes |
| 108 | Per1 | 10444 | -0.483 | -0.0977 | Yes |
| 109 | Tsc22d1 | 10458 | -0.486 | -0.0848 | Yes |
| 110 | Klf9 | 10732 | -0.541 | -0.0930 | Yes |
| 111 | Plaur | 10911 | -0.580 | -0.0918 | Yes |
| 112 | Abca1 | 11001 | -0.606 | -0.0820 | Yes |
| 113 | Bcl6 | 11076 | -0.631 | -0.0703 | Yes |
| 114 | Il6st | 11110 | -0.641 | -0.0547 | Yes |
| 115 | Pdlim5 | 11277 | -0.718 | -0.0484 | Yes |
| 116 | Ets2 | 11337 | -0.748 | -0.0320 | Yes |
| 117 | Sod2 | 11509 | -0.912 | -0.0206 | Yes |
| 118 | Ifngr2 | 11552 | -1.012 | 0.0050 | Yes |