Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
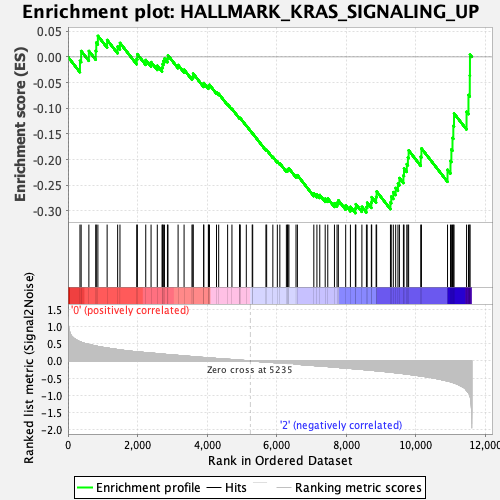
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
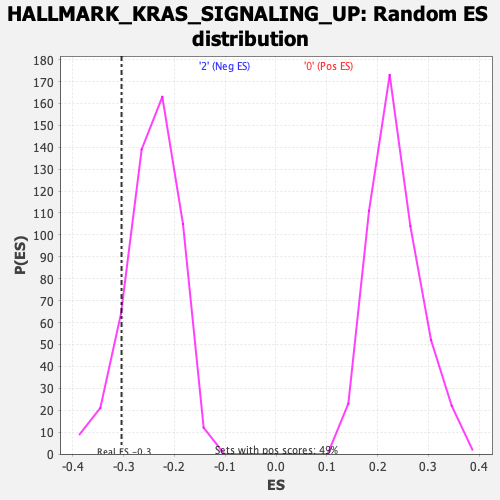
| Enrichment Score (ES) | -0.30427685 |
| Normalized Enrichment Score (NES) | -1.2582184 |
| Nominal p-value | 0.08966862 |
| FDR q-value | 0.389863 |
| FWER p-Value | 0.816 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tspan13 | 344 | 0.556 | -0.0074 | No |
| 2 | Wdr33 | 378 | 0.540 | 0.0116 | No |
| 3 | Tnfaip3 | 600 | 0.476 | 0.0116 | No |
| 4 | Il1rl2 | 798 | 0.430 | 0.0119 | No |
| 5 | Laptm5 | 813 | 0.428 | 0.0280 | No |
| 6 | Yrdc | 859 | 0.419 | 0.0410 | No |
| 7 | Ero1a | 1125 | 0.373 | 0.0331 | No |
| 8 | Adam8 | 1427 | 0.328 | 0.0202 | No |
| 9 | Map7 | 1494 | 0.319 | 0.0273 | No |
| 10 | Ppp1r15a | 1976 | 0.263 | -0.0038 | No |
| 11 | Scg5 | 1994 | 0.261 | 0.0053 | No |
| 12 | Btbd3 | 2235 | 0.239 | -0.0059 | No |
| 13 | Tfpi | 2388 | 0.225 | -0.0100 | No |
| 14 | Prelid3b | 2567 | 0.210 | -0.0170 | No |
| 15 | Ccser2 | 2704 | 0.198 | -0.0208 | No |
| 16 | Trib1 | 2719 | 0.196 | -0.0141 | No |
| 17 | Car2 | 2741 | 0.195 | -0.0080 | No |
| 18 | Ano1 | 2773 | 0.191 | -0.0030 | No |
| 19 | Zfp639 | 2859 | 0.183 | -0.0030 | No |
| 20 | Cbx8 | 2874 | 0.182 | 0.0031 | No |
| 21 | Cab39l | 3165 | 0.158 | -0.0157 | No |
| 22 | Psmb8 | 3337 | 0.142 | -0.0248 | No |
| 23 | Adgrl4 | 3564 | 0.124 | -0.0394 | No |
| 24 | Bpgm | 3594 | 0.120 | -0.0370 | No |
| 25 | St6gal1 | 3596 | 0.120 | -0.0323 | No |
| 26 | Fbxo4 | 3900 | 0.098 | -0.0546 | No |
| 27 | Lcp1 | 3903 | 0.098 | -0.0508 | No |
| 28 | Strn | 4035 | 0.088 | -0.0586 | No |
| 29 | Glrx | 4043 | 0.087 | -0.0557 | No |
| 30 | Nin | 4067 | 0.086 | -0.0542 | No |
| 31 | Cbr4 | 4269 | 0.069 | -0.0689 | No |
| 32 | Adam17 | 4332 | 0.065 | -0.0717 | No |
| 33 | Dock2 | 4589 | 0.047 | -0.0920 | No |
| 34 | Plvap | 4713 | 0.038 | -0.1011 | No |
| 35 | F13a1 | 4932 | 0.022 | -0.1192 | No |
| 36 | Tnfrsf1b | 4938 | 0.021 | -0.1188 | No |
| 37 | Map4k1 | 4950 | 0.020 | -0.1189 | No |
| 38 | H2bc3 | 5126 | 0.008 | -0.1338 | No |
| 39 | Mtmr10 | 5296 | -0.004 | -0.1483 | No |
| 40 | Cxcr4 | 5310 | -0.005 | -0.1492 | No |
| 41 | Birc3 | 5690 | -0.032 | -0.1809 | No |
| 42 | Tmem176b | 5711 | -0.034 | -0.1812 | No |
| 43 | Crot | 5889 | -0.046 | -0.1947 | No |
| 44 | Map3k1 | 6018 | -0.055 | -0.2036 | No |
| 45 | Ccnd2 | 6090 | -0.060 | -0.2073 | No |
| 46 | Fuca1 | 6278 | -0.075 | -0.2206 | No |
| 47 | Ets1 | 6311 | -0.077 | -0.2202 | No |
| 48 | Angptl4 | 6325 | -0.078 | -0.2182 | No |
| 49 | Ikzf1 | 6351 | -0.080 | -0.2171 | No |
| 50 | Irf8 | 6555 | -0.094 | -0.2310 | No |
| 51 | Cbl | 6595 | -0.098 | -0.2304 | No |
| 52 | Jup | 7067 | -0.134 | -0.2659 | No |
| 53 | Mmd | 7156 | -0.141 | -0.2678 | No |
| 54 | Pecam1 | 7238 | -0.147 | -0.2689 | No |
| 55 | Hdac9 | 7394 | -0.158 | -0.2760 | No |
| 56 | Cd37 | 7468 | -0.163 | -0.2757 | No |
| 57 | Pdcd1lg2 | 7660 | -0.181 | -0.2850 | No |
| 58 | Itga2 | 7735 | -0.187 | -0.2839 | No |
| 59 | Satb1 | 7772 | -0.190 | -0.2793 | No |
| 60 | Eng | 7983 | -0.209 | -0.2891 | No |
| 61 | Akt2 | 8119 | -0.219 | -0.2919 | No |
| 62 | Spry2 | 8262 | -0.230 | -0.2950 | Yes |
| 63 | Tmem176a | 8275 | -0.231 | -0.2867 | Yes |
| 64 | Atg10 | 8449 | -0.246 | -0.2918 | Yes |
| 65 | Ank | 8579 | -0.259 | -0.2925 | Yes |
| 66 | Dusp6 | 8600 | -0.260 | -0.2837 | Yes |
| 67 | Ptbp2 | 8721 | -0.270 | -0.2833 | Yes |
| 68 | Dnmbp | 8731 | -0.272 | -0.2731 | Yes |
| 69 | Etv5 | 8858 | -0.285 | -0.2725 | Yes |
| 70 | Fcer1g | 8873 | -0.287 | -0.2621 | Yes |
| 71 | Galnt3 | 9270 | -0.328 | -0.2832 | Yes |
| 72 | Csf2ra | 9293 | -0.331 | -0.2718 | Yes |
| 73 | Sdccag8 | 9352 | -0.335 | -0.2633 | Yes |
| 74 | Nrp1 | 9420 | -0.344 | -0.2552 | Yes |
| 75 | Ammecr1 | 9486 | -0.351 | -0.2466 | Yes |
| 76 | Evi5 | 9528 | -0.355 | -0.2358 | Yes |
| 77 | Zfp277 | 9643 | -0.369 | -0.2308 | Yes |
| 78 | Avl9 | 9660 | -0.371 | -0.2172 | Yes |
| 79 | Vwa5a | 9740 | -0.380 | -0.2087 | Yes |
| 80 | Kcnn4 | 9772 | -0.383 | -0.1959 | Yes |
| 81 | Itgb2 | 9793 | -0.387 | -0.1820 | Yes |
| 82 | Tor1aip2 | 10141 | -0.440 | -0.1944 | Yes |
| 83 | Gypc | 10161 | -0.442 | -0.1782 | Yes |
| 84 | Plaur | 10911 | -0.580 | -0.2198 | Yes |
| 85 | Lat2 | 10996 | -0.606 | -0.2026 | Yes |
| 86 | Ptcd2 | 11022 | -0.613 | -0.1800 | Yes |
| 87 | Rabgap1l | 11054 | -0.620 | -0.1577 | Yes |
| 88 | Hsd11b1 | 11078 | -0.631 | -0.1341 | Yes |
| 89 | Cdadc1 | 11098 | -0.636 | -0.1101 | Yes |
| 90 | Cmklr1 | 11458 | -0.854 | -0.1068 | Yes |
| 91 | Rbm4 | 11513 | -0.925 | -0.0741 | Yes |
| 92 | Il2rg | 11551 | -1.012 | -0.0364 | Yes |
| 93 | Usp12 | 11556 | -1.023 | 0.0046 | Yes |