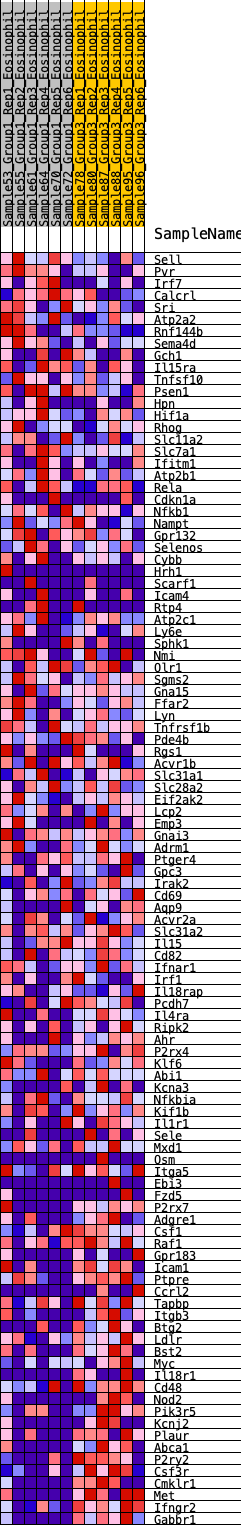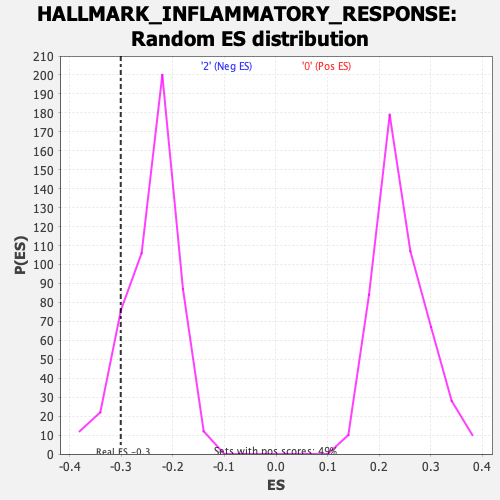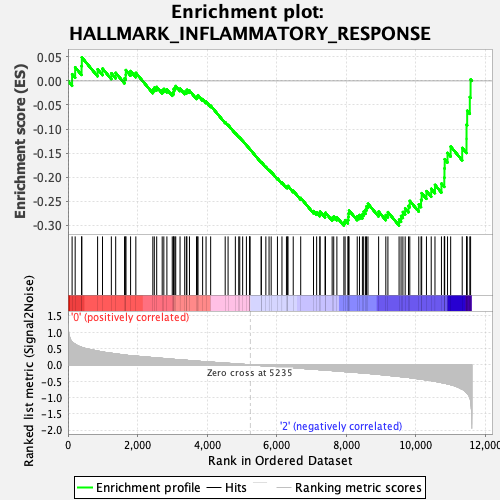
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.30089265 |
| Normalized Enrichment Score (NES) | -1.2527854 |
| Nominal p-value | 0.10873786 |
| FDR q-value | 0.36966032 |
| FWER p-Value | 0.828 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sell | 118 | 0.702 | 0.0141 | No |
| 2 | Pvr | 206 | 0.630 | 0.0283 | No |
| 3 | Irf7 | 390 | 0.536 | 0.0310 | No |
| 4 | Calcrl | 398 | 0.533 | 0.0489 | No |
| 5 | Sri | 851 | 0.420 | 0.0242 | No |
| 6 | Atp2a2 | 992 | 0.395 | 0.0257 | No |
| 7 | Rnf144b | 1246 | 0.354 | 0.0160 | No |
| 8 | Sema4d | 1371 | 0.336 | 0.0168 | No |
| 9 | Gch1 | 1625 | 0.301 | 0.0053 | No |
| 10 | Il15ra | 1660 | 0.296 | 0.0126 | No |
| 11 | Tnfsf10 | 1663 | 0.296 | 0.0227 | No |
| 12 | Psen1 | 1799 | 0.280 | 0.0206 | No |
| 13 | Hpn | 1950 | 0.266 | 0.0168 | No |
| 14 | Hif1a | 2439 | 0.221 | -0.0179 | No |
| 15 | Rhog | 2481 | 0.218 | -0.0139 | No |
| 16 | Slc11a2 | 2550 | 0.213 | -0.0125 | No |
| 17 | Slc7a1 | 2708 | 0.197 | -0.0193 | No |
| 18 | Ifitm1 | 2755 | 0.193 | -0.0166 | No |
| 19 | Atp2b1 | 2843 | 0.184 | -0.0178 | No |
| 20 | Rela | 2997 | 0.171 | -0.0251 | No |
| 21 | Cdkn1a | 3035 | 0.167 | -0.0225 | No |
| 22 | Nfkb1 | 3038 | 0.167 | -0.0169 | No |
| 23 | Nampt | 3072 | 0.165 | -0.0141 | No |
| 24 | Gpr132 | 3097 | 0.163 | -0.0105 | No |
| 25 | Selenos | 3221 | 0.153 | -0.0159 | No |
| 26 | Cybb | 3355 | 0.141 | -0.0226 | No |
| 27 | Hrh1 | 3403 | 0.138 | -0.0219 | No |
| 28 | Scarf1 | 3418 | 0.136 | -0.0184 | No |
| 29 | Icam4 | 3492 | 0.130 | -0.0202 | No |
| 30 | Rtp4 | 3694 | 0.113 | -0.0338 | No |
| 31 | Atp2c1 | 3718 | 0.111 | -0.0319 | No |
| 32 | Ly6e | 3738 | 0.110 | -0.0298 | No |
| 33 | Sphk1 | 3870 | 0.100 | -0.0377 | No |
| 34 | Nmi | 3969 | 0.093 | -0.0430 | No |
| 35 | Olr1 | 4101 | 0.083 | -0.0515 | No |
| 36 | Sgms2 | 4519 | 0.051 | -0.0860 | No |
| 37 | Gna15 | 4602 | 0.046 | -0.0915 | No |
| 38 | Ffar2 | 4811 | 0.032 | -0.1085 | No |
| 39 | Lyn | 4909 | 0.024 | -0.1161 | No |
| 40 | Tnfrsf1b | 4938 | 0.021 | -0.1178 | No |
| 41 | Pde4b | 5021 | 0.015 | -0.1244 | No |
| 42 | Rgs1 | 5129 | 0.008 | -0.1334 | No |
| 43 | Acvr1b | 5222 | 0.001 | -0.1414 | No |
| 44 | Slc31a1 | 5224 | 0.001 | -0.1415 | No |
| 45 | Slc28a2 | 5231 | 0.000 | -0.1420 | No |
| 46 | Eif2ak2 | 5554 | -0.023 | -0.1692 | No |
| 47 | Lcp2 | 5557 | -0.023 | -0.1686 | No |
| 48 | Emp3 | 5688 | -0.032 | -0.1788 | No |
| 49 | Gnai3 | 5780 | -0.039 | -0.1853 | No |
| 50 | Adrm1 | 5841 | -0.043 | -0.1891 | No |
| 51 | Ptger4 | 6021 | -0.056 | -0.2027 | No |
| 52 | Gpc3 | 6151 | -0.065 | -0.2117 | No |
| 53 | Irak2 | 6280 | -0.075 | -0.2202 | No |
| 54 | Cd69 | 6314 | -0.077 | -0.2204 | No |
| 55 | Aqp9 | 6323 | -0.078 | -0.2184 | No |
| 56 | Acvr2a | 6475 | -0.090 | -0.2284 | No |
| 57 | Slc31a2 | 6690 | -0.105 | -0.2433 | No |
| 58 | Il15 | 7058 | -0.133 | -0.2706 | No |
| 59 | Cd82 | 7148 | -0.140 | -0.2735 | No |
| 60 | Ifnar1 | 7240 | -0.147 | -0.2763 | No |
| 61 | Irf1 | 7246 | -0.148 | -0.2716 | No |
| 62 | Il18rap | 7390 | -0.158 | -0.2786 | No |
| 63 | Pcdh7 | 7403 | -0.159 | -0.2741 | No |
| 64 | Il4ra | 7594 | -0.175 | -0.2845 | No |
| 65 | Ripk2 | 7639 | -0.179 | -0.2821 | No |
| 66 | Ahr | 7730 | -0.186 | -0.2835 | No |
| 67 | P2rx4 | 7931 | -0.204 | -0.2938 | Yes |
| 68 | Klf6 | 7966 | -0.208 | -0.2896 | Yes |
| 69 | Abi1 | 8047 | -0.213 | -0.2891 | Yes |
| 70 | Kcna3 | 8060 | -0.214 | -0.2828 | Yes |
| 71 | Nfkbia | 8065 | -0.215 | -0.2756 | Yes |
| 72 | Kif1b | 8080 | -0.216 | -0.2694 | Yes |
| 73 | Il1r1 | 8315 | -0.235 | -0.2816 | Yes |
| 74 | Sele | 8380 | -0.240 | -0.2788 | Yes |
| 75 | Mxd1 | 8464 | -0.248 | -0.2775 | Yes |
| 76 | Osm | 8500 | -0.251 | -0.2718 | Yes |
| 77 | Itga5 | 8555 | -0.256 | -0.2676 | Yes |
| 78 | Ebi3 | 8581 | -0.259 | -0.2608 | Yes |
| 79 | Fzd5 | 8623 | -0.262 | -0.2553 | Yes |
| 80 | P2rx7 | 8930 | -0.293 | -0.2718 | Yes |
| 81 | Adgre1 | 9137 | -0.313 | -0.2788 | Yes |
| 82 | Csf1 | 9197 | -0.320 | -0.2729 | Yes |
| 83 | Raf1 | 9518 | -0.354 | -0.2884 | Yes |
| 84 | Gpr183 | 9575 | -0.362 | -0.2807 | Yes |
| 85 | Icam1 | 9628 | -0.367 | -0.2725 | Yes |
| 86 | Ptpre | 9693 | -0.375 | -0.2651 | Yes |
| 87 | Ccrl2 | 9789 | -0.387 | -0.2599 | Yes |
| 88 | Tapbp | 9826 | -0.392 | -0.2495 | Yes |
| 89 | Itgb3 | 10084 | -0.430 | -0.2569 | Yes |
| 90 | Btg2 | 10150 | -0.441 | -0.2473 | Yes |
| 91 | Ldlr | 10168 | -0.443 | -0.2334 | Yes |
| 92 | Bst2 | 10305 | -0.462 | -0.2292 | Yes |
| 93 | Myc | 10442 | -0.482 | -0.2243 | Yes |
| 94 | Il18r1 | 10550 | -0.504 | -0.2161 | Yes |
| 95 | Cd48 | 10736 | -0.542 | -0.2134 | Yes |
| 96 | Nod2 | 10818 | -0.561 | -0.2010 | Yes |
| 97 | Pik3r5 | 10823 | -0.562 | -0.1818 | Yes |
| 98 | Kcnj2 | 10830 | -0.563 | -0.1628 | Yes |
| 99 | Plaur | 10911 | -0.580 | -0.1497 | Yes |
| 100 | Abca1 | 11001 | -0.606 | -0.1364 | Yes |
| 101 | P2ry2 | 11333 | -0.746 | -0.1393 | Yes |
| 102 | Csf3r | 11457 | -0.853 | -0.1204 | Yes |
| 103 | Cmklr1 | 11458 | -0.854 | -0.0908 | Yes |
| 104 | Met | 11476 | -0.876 | -0.0620 | Yes |
| 105 | Ifngr2 | 11552 | -1.012 | -0.0334 | Yes |
| 106 | Gabbr1 | 11574 | -1.104 | 0.0030 | Yes |