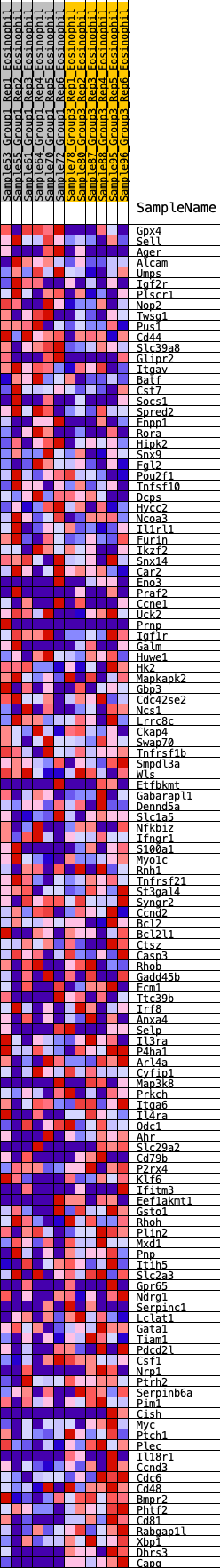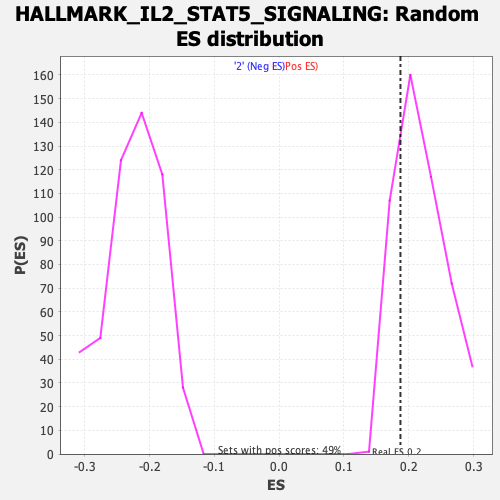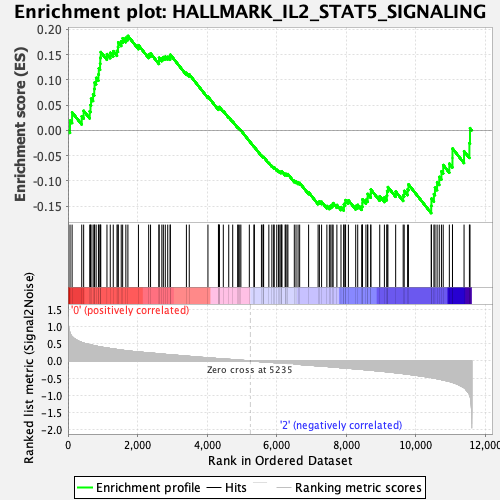
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_IL2_STAT5_SIGNALING |
| Enrichment Score (ES) | 0.18713985 |
| Normalized Enrichment Score (NES) | 0.85167205 |
| Nominal p-value | 0.77732795 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx4 | 58 | 0.811 | 0.0196 | Yes |
| 2 | Sell | 118 | 0.702 | 0.0357 | Yes |
| 3 | Ager | 395 | 0.535 | 0.0279 | Yes |
| 4 | Alcam | 450 | 0.514 | 0.0388 | Yes |
| 5 | Umps | 625 | 0.470 | 0.0379 | Yes |
| 6 | Igf2r | 647 | 0.466 | 0.0503 | Yes |
| 7 | Plscr1 | 663 | 0.463 | 0.0630 | Yes |
| 8 | Nop2 | 721 | 0.448 | 0.0716 | Yes |
| 9 | Twsg1 | 754 | 0.443 | 0.0823 | Yes |
| 10 | Pus1 | 764 | 0.441 | 0.0949 | Yes |
| 11 | Cd44 | 809 | 0.429 | 0.1041 | Yes |
| 12 | Slc39a8 | 875 | 0.416 | 0.1110 | Yes |
| 13 | Glipr2 | 886 | 0.414 | 0.1227 | Yes |
| 14 | Itgav | 922 | 0.408 | 0.1321 | Yes |
| 15 | Batf | 928 | 0.407 | 0.1440 | Yes |
| 16 | Cst7 | 942 | 0.405 | 0.1552 | Yes |
| 17 | Socs1 | 1120 | 0.373 | 0.1511 | Yes |
| 18 | Spred2 | 1215 | 0.359 | 0.1538 | Yes |
| 19 | Enpp1 | 1301 | 0.346 | 0.1569 | Yes |
| 20 | Rora | 1409 | 0.330 | 0.1575 | Yes |
| 21 | Hipk2 | 1437 | 0.326 | 0.1651 | Yes |
| 22 | Snx9 | 1444 | 0.324 | 0.1744 | Yes |
| 23 | Fgl2 | 1530 | 0.315 | 0.1765 | Yes |
| 24 | Pou2f1 | 1571 | 0.308 | 0.1824 | Yes |
| 25 | Tnfsf10 | 1663 | 0.296 | 0.1835 | Yes |
| 26 | Dcps | 1722 | 0.288 | 0.1871 | Yes |
| 27 | Hycc2 | 2025 | 0.259 | 0.1687 | No |
| 28 | Ncoa3 | 2319 | 0.232 | 0.1502 | No |
| 29 | Il1rl1 | 2373 | 0.227 | 0.1525 | No |
| 30 | Furin | 2616 | 0.206 | 0.1377 | No |
| 31 | Ikzf2 | 2617 | 0.206 | 0.1439 | No |
| 32 | Snx14 | 2698 | 0.199 | 0.1430 | No |
| 33 | Car2 | 2741 | 0.195 | 0.1452 | No |
| 34 | Eno3 | 2797 | 0.189 | 0.1462 | No |
| 35 | Praf2 | 2867 | 0.182 | 0.1457 | No |
| 36 | Ccne1 | 2936 | 0.175 | 0.1451 | No |
| 37 | Uck2 | 2942 | 0.175 | 0.1500 | No |
| 38 | Prnp | 3404 | 0.138 | 0.1140 | No |
| 39 | Igf1r | 3486 | 0.130 | 0.1109 | No |
| 40 | Galm | 4023 | 0.089 | 0.0669 | No |
| 41 | Huwe1 | 4325 | 0.065 | 0.0427 | No |
| 42 | Hk2 | 4327 | 0.065 | 0.0446 | No |
| 43 | Mapkapk2 | 4347 | 0.064 | 0.0448 | No |
| 44 | Gbp3 | 4351 | 0.063 | 0.0465 | No |
| 45 | Cdc42se2 | 4465 | 0.055 | 0.0383 | No |
| 46 | Ncs1 | 4621 | 0.044 | 0.0262 | No |
| 47 | Lrrc8c | 4735 | 0.037 | 0.0175 | No |
| 48 | Ckap4 | 4878 | 0.026 | 0.0059 | No |
| 49 | Swap70 | 4897 | 0.025 | 0.0051 | No |
| 50 | Tnfrsf1b | 4938 | 0.021 | 0.0022 | No |
| 51 | Smpdl3a | 4970 | 0.019 | 0.0001 | No |
| 52 | Wls | 5213 | 0.001 | -0.0209 | No |
| 53 | Etfbkmt | 5344 | -0.008 | -0.0320 | No |
| 54 | Gabarapl1 | 5359 | -0.008 | -0.0330 | No |
| 55 | Dennd5a | 5561 | -0.023 | -0.0498 | No |
| 56 | Slc1a5 | 5606 | -0.027 | -0.0528 | No |
| 57 | Nfkbiz | 5616 | -0.027 | -0.0528 | No |
| 58 | Ifngr1 | 5624 | -0.028 | -0.0525 | No |
| 59 | S100a1 | 5774 | -0.038 | -0.0643 | No |
| 60 | Myo1c | 5861 | -0.044 | -0.0705 | No |
| 61 | Rnh1 | 5913 | -0.048 | -0.0735 | No |
| 62 | Tnfrsf21 | 5927 | -0.049 | -0.0731 | No |
| 63 | St3gal4 | 6001 | -0.055 | -0.0778 | No |
| 64 | Syngr2 | 6050 | -0.058 | -0.0802 | No |
| 65 | Ccnd2 | 6090 | -0.060 | -0.0818 | No |
| 66 | Bcl2 | 6123 | -0.063 | -0.0827 | No |
| 67 | Bcl2l1 | 6142 | -0.064 | -0.0823 | No |
| 68 | Ctsz | 6149 | -0.064 | -0.0809 | No |
| 69 | Casp3 | 6242 | -0.072 | -0.0867 | No |
| 70 | Rhob | 6259 | -0.073 | -0.0859 | No |
| 71 | Gadd45b | 6295 | -0.076 | -0.0866 | No |
| 72 | Ecm1 | 6322 | -0.078 | -0.0865 | No |
| 73 | Ttc39b | 6506 | -0.091 | -0.0997 | No |
| 74 | Irf8 | 6555 | -0.094 | -0.1010 | No |
| 75 | Anxa4 | 6614 | -0.099 | -0.1030 | No |
| 76 | Selp | 6660 | -0.102 | -0.1039 | No |
| 77 | Il3ra | 6917 | -0.122 | -0.1225 | No |
| 78 | P4ha1 | 7190 | -0.144 | -0.1418 | No |
| 79 | Arl4a | 7227 | -0.147 | -0.1405 | No |
| 80 | Cyfip1 | 7290 | -0.151 | -0.1413 | No |
| 81 | Map3k8 | 7442 | -0.161 | -0.1495 | No |
| 82 | Prkch | 7514 | -0.168 | -0.1506 | No |
| 83 | Itga6 | 7555 | -0.171 | -0.1489 | No |
| 84 | Il4ra | 7594 | -0.175 | -0.1469 | No |
| 85 | Odc1 | 7622 | -0.178 | -0.1439 | No |
| 86 | Ahr | 7730 | -0.186 | -0.1475 | No |
| 87 | Slc29a2 | 7848 | -0.197 | -0.1517 | No |
| 88 | Cd79b | 7928 | -0.204 | -0.1524 | No |
| 89 | P2rx4 | 7931 | -0.204 | -0.1464 | No |
| 90 | Klf6 | 7966 | -0.208 | -0.1431 | No |
| 91 | Ifitm3 | 7980 | -0.209 | -0.1379 | No |
| 92 | Eef1akmt1 | 8061 | -0.214 | -0.1383 | No |
| 93 | Gsto1 | 8271 | -0.230 | -0.1495 | No |
| 94 | Rhoh | 8329 | -0.236 | -0.1473 | No |
| 95 | Plin2 | 8444 | -0.246 | -0.1498 | No |
| 96 | Mxd1 | 8464 | -0.248 | -0.1439 | No |
| 97 | Pnp | 8465 | -0.248 | -0.1364 | No |
| 98 | Itih5 | 8565 | -0.257 | -0.1372 | No |
| 99 | Slc2a3 | 8615 | -0.261 | -0.1336 | No |
| 100 | Gpr65 | 8616 | -0.261 | -0.1256 | No |
| 101 | Ndrg1 | 8704 | -0.269 | -0.1251 | No |
| 102 | Serpinc1 | 8706 | -0.269 | -0.1170 | No |
| 103 | Lclat1 | 8962 | -0.295 | -0.1302 | No |
| 104 | Gata1 | 9096 | -0.308 | -0.1325 | No |
| 105 | Tiam1 | 9166 | -0.316 | -0.1289 | No |
| 106 | Pdcd2l | 9174 | -0.317 | -0.1199 | No |
| 107 | Csf1 | 9197 | -0.320 | -0.1121 | No |
| 108 | Nrp1 | 9420 | -0.344 | -0.1210 | No |
| 109 | Ptrh2 | 9636 | -0.369 | -0.1285 | No |
| 110 | Serpinb6a | 9665 | -0.372 | -0.1196 | No |
| 111 | Pim1 | 9763 | -0.382 | -0.1165 | No |
| 112 | Cish | 9787 | -0.386 | -0.1068 | No |
| 113 | Myc | 10442 | -0.482 | -0.1491 | No |
| 114 | Ptch1 | 10449 | -0.484 | -0.1349 | No |
| 115 | Plec | 10520 | -0.497 | -0.1259 | No |
| 116 | Il18r1 | 10550 | -0.504 | -0.1132 | No |
| 117 | Ccnd3 | 10611 | -0.517 | -0.1027 | No |
| 118 | Cdc6 | 10673 | -0.530 | -0.0920 | No |
| 119 | Cd48 | 10736 | -0.542 | -0.0809 | No |
| 120 | Bmpr2 | 10787 | -0.555 | -0.0684 | No |
| 121 | Phtf2 | 10963 | -0.592 | -0.0657 | No |
| 122 | Cd81 | 11053 | -0.620 | -0.0546 | No |
| 123 | Rabgap1l | 11054 | -0.620 | -0.0358 | No |
| 124 | Xbp1 | 11386 | -0.774 | -0.0412 | No |
| 125 | Dhrs3 | 11540 | -0.966 | -0.0252 | No |
| 126 | Capg | 11557 | -1.025 | 0.0045 | No |