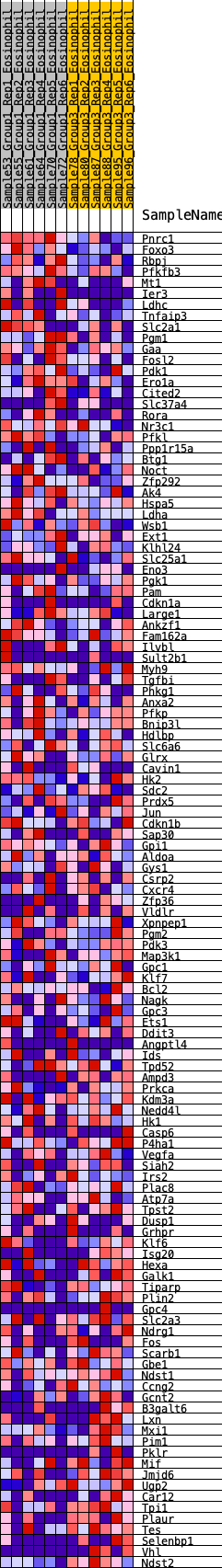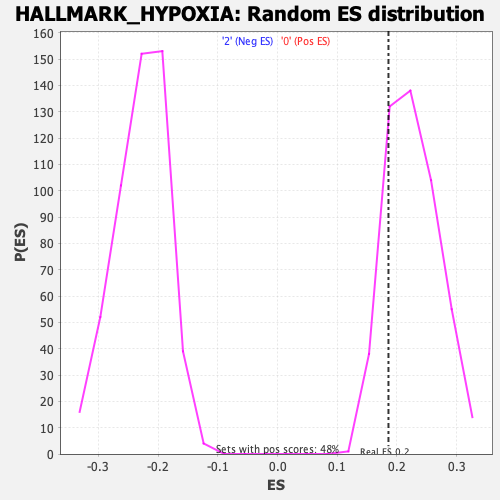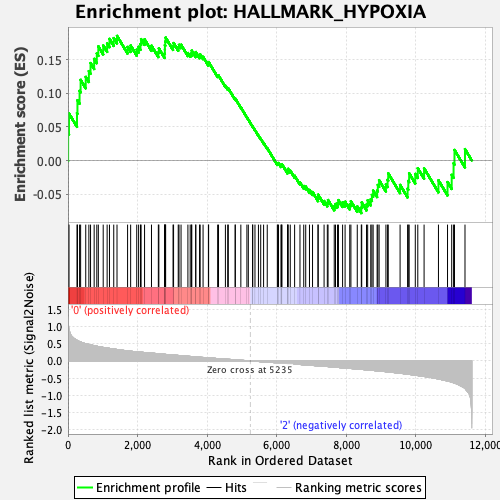
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.18584391 |
| Normalized Enrichment Score (NES) | 0.8235886 |
| Nominal p-value | 0.8755187 |
| FDR q-value | 0.9367763 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pnrc1 | 4 | 1.194 | 0.0407 | Yes |
| 2 | Foxo3 | 29 | 0.914 | 0.0701 | Yes |
| 3 | Rbpj | 259 | 0.596 | 0.0707 | Yes |
| 4 | Pfkfb3 | 272 | 0.588 | 0.0899 | Yes |
| 5 | Mt1 | 333 | 0.560 | 0.1039 | Yes |
| 6 | Ier3 | 363 | 0.550 | 0.1203 | Yes |
| 7 | Ldhc | 510 | 0.497 | 0.1247 | Yes |
| 8 | Tnfaip3 | 600 | 0.476 | 0.1334 | Yes |
| 9 | Slc2a1 | 648 | 0.466 | 0.1453 | Yes |
| 10 | Pgm1 | 752 | 0.443 | 0.1516 | Yes |
| 11 | Gaa | 828 | 0.424 | 0.1597 | Yes |
| 12 | Fosl2 | 872 | 0.417 | 0.1703 | Yes |
| 13 | Pdk1 | 1012 | 0.391 | 0.1716 | Yes |
| 14 | Ero1a | 1125 | 0.373 | 0.1747 | Yes |
| 15 | Cited2 | 1192 | 0.362 | 0.1814 | Yes |
| 16 | Slc37a4 | 1315 | 0.344 | 0.1826 | Yes |
| 17 | Rora | 1409 | 0.330 | 0.1858 | Yes |
| 18 | Nr3c1 | 1713 | 0.288 | 0.1694 | No |
| 19 | Pfkl | 1801 | 0.280 | 0.1714 | No |
| 20 | Ppp1r15a | 1976 | 0.263 | 0.1653 | No |
| 21 | Btg1 | 2030 | 0.258 | 0.1696 | No |
| 22 | Noct | 2084 | 0.253 | 0.1737 | No |
| 23 | Zfp292 | 2101 | 0.252 | 0.1810 | No |
| 24 | Ak4 | 2201 | 0.242 | 0.1807 | No |
| 25 | Hspa5 | 2400 | 0.224 | 0.1712 | No |
| 26 | Ldha | 2597 | 0.208 | 0.1613 | No |
| 27 | Wsb1 | 2612 | 0.207 | 0.1672 | No |
| 28 | Ext1 | 2777 | 0.191 | 0.1595 | No |
| 29 | Klhl24 | 2784 | 0.190 | 0.1655 | No |
| 30 | Slc25a1 | 2786 | 0.190 | 0.1720 | No |
| 31 | Eno3 | 2797 | 0.189 | 0.1776 | No |
| 32 | Pgk1 | 2807 | 0.188 | 0.1833 | No |
| 33 | Pam | 3020 | 0.169 | 0.1706 | No |
| 34 | Cdkn1a | 3035 | 0.167 | 0.1752 | No |
| 35 | Large1 | 3169 | 0.158 | 0.1690 | No |
| 36 | Ankzf1 | 3191 | 0.156 | 0.1726 | No |
| 37 | Fam162a | 3250 | 0.150 | 0.1727 | No |
| 38 | Ilvbl | 3447 | 0.134 | 0.1602 | No |
| 39 | Sult2b1 | 3513 | 0.129 | 0.1590 | No |
| 40 | Myh9 | 3553 | 0.125 | 0.1599 | No |
| 41 | Tgfbi | 3557 | 0.124 | 0.1639 | No |
| 42 | Phkg1 | 3671 | 0.115 | 0.1580 | No |
| 43 | Anxa2 | 3674 | 0.115 | 0.1618 | No |
| 44 | Pfkp | 3782 | 0.107 | 0.1562 | No |
| 45 | Bnip3l | 3799 | 0.105 | 0.1584 | No |
| 46 | Hdlbp | 3883 | 0.100 | 0.1546 | No |
| 47 | Slc6a6 | 4036 | 0.088 | 0.1444 | No |
| 48 | Glrx | 4043 | 0.087 | 0.1469 | No |
| 49 | Cavin1 | 4302 | 0.067 | 0.1267 | No |
| 50 | Hk2 | 4327 | 0.065 | 0.1269 | No |
| 51 | Sdc2 | 4525 | 0.051 | 0.1114 | No |
| 52 | Prdx5 | 4592 | 0.046 | 0.1073 | No |
| 53 | Jun | 4608 | 0.045 | 0.1076 | No |
| 54 | Cdkn1b | 4805 | 0.032 | 0.0916 | No |
| 55 | Sap30 | 4810 | 0.032 | 0.0923 | No |
| 56 | Gpi1 | 4969 | 0.019 | 0.0792 | No |
| 57 | Aldoa | 5143 | 0.006 | 0.0644 | No |
| 58 | Gys1 | 5184 | 0.003 | 0.0610 | No |
| 59 | Csrp2 | 5307 | -0.005 | 0.0506 | No |
| 60 | Cxcr4 | 5310 | -0.005 | 0.0506 | No |
| 61 | Zfp36 | 5375 | -0.009 | 0.0453 | No |
| 62 | Vldlr | 5475 | -0.017 | 0.0373 | No |
| 63 | Xpnpep1 | 5540 | -0.022 | 0.0325 | No |
| 64 | Pgm2 | 5623 | -0.028 | 0.0263 | No |
| 65 | Pdk3 | 5728 | -0.035 | 0.0184 | No |
| 66 | Map3k1 | 6018 | -0.055 | -0.0048 | No |
| 67 | Gpc1 | 6030 | -0.056 | -0.0038 | No |
| 68 | Klf7 | 6052 | -0.058 | -0.0037 | No |
| 69 | Bcl2 | 6123 | -0.063 | -0.0076 | No |
| 70 | Nagk | 6137 | -0.064 | -0.0065 | No |
| 71 | Gpc3 | 6151 | -0.065 | -0.0055 | No |
| 72 | Ets1 | 6311 | -0.077 | -0.0166 | No |
| 73 | Ddit3 | 6324 | -0.078 | -0.0150 | No |
| 74 | Angptl4 | 6325 | -0.078 | -0.0123 | No |
| 75 | Ids | 6390 | -0.083 | -0.0151 | No |
| 76 | Tpd52 | 6515 | -0.092 | -0.0227 | No |
| 77 | Ampd3 | 6669 | -0.103 | -0.0325 | No |
| 78 | Prkca | 6777 | -0.112 | -0.0379 | No |
| 79 | Kdm3a | 6834 | -0.117 | -0.0388 | No |
| 80 | Nedd4l | 6942 | -0.124 | -0.0438 | No |
| 81 | Hk1 | 7030 | -0.130 | -0.0469 | No |
| 82 | Casp6 | 7189 | -0.144 | -0.0557 | No |
| 83 | P4ha1 | 7190 | -0.144 | -0.0508 | No |
| 84 | Vegfa | 7362 | -0.156 | -0.0603 | No |
| 85 | Siah2 | 7455 | -0.162 | -0.0627 | No |
| 86 | Irs2 | 7476 | -0.164 | -0.0589 | No |
| 87 | Plac8 | 7654 | -0.180 | -0.0680 | No |
| 88 | Atp7a | 7690 | -0.183 | -0.0648 | No |
| 89 | Tpst2 | 7749 | -0.188 | -0.0634 | No |
| 90 | Dusp1 | 7775 | -0.191 | -0.0590 | No |
| 91 | Grhpr | 7894 | -0.201 | -0.0623 | No |
| 92 | Klf6 | 7966 | -0.208 | -0.0614 | No |
| 93 | Isg20 | 8100 | -0.218 | -0.0654 | No |
| 94 | Hexa | 8133 | -0.220 | -0.0607 | No |
| 95 | Galk1 | 8316 | -0.235 | -0.0684 | No |
| 96 | Tiparp | 8432 | -0.244 | -0.0700 | No |
| 97 | Plin2 | 8444 | -0.246 | -0.0625 | No |
| 98 | Gpc4 | 8582 | -0.259 | -0.0655 | No |
| 99 | Slc2a3 | 8615 | -0.261 | -0.0593 | No |
| 100 | Ndrg1 | 8704 | -0.269 | -0.0577 | No |
| 101 | Fos | 8737 | -0.272 | -0.0511 | No |
| 102 | Scarb1 | 8772 | -0.275 | -0.0446 | No |
| 103 | Gbe1 | 8887 | -0.288 | -0.0446 | No |
| 104 | Ndst1 | 8907 | -0.290 | -0.0363 | No |
| 105 | Ccng2 | 8944 | -0.294 | -0.0293 | No |
| 106 | Gcnt2 | 9140 | -0.313 | -0.0355 | No |
| 107 | B3galt6 | 9188 | -0.319 | -0.0286 | No |
| 108 | Lxn | 9206 | -0.321 | -0.0191 | No |
| 109 | Mxi1 | 9547 | -0.359 | -0.0363 | No |
| 110 | Pim1 | 9763 | -0.382 | -0.0419 | No |
| 111 | Pklr | 9783 | -0.386 | -0.0303 | No |
| 112 | Mif | 9808 | -0.388 | -0.0190 | No |
| 113 | Jmjd6 | 9984 | -0.416 | -0.0199 | No |
| 114 | Ugp2 | 10056 | -0.425 | -0.0115 | No |
| 115 | Car12 | 10237 | -0.452 | -0.0116 | No |
| 116 | Tpi1 | 10650 | -0.524 | -0.0294 | No |
| 117 | Plaur | 10911 | -0.580 | -0.0321 | No |
| 118 | Tes | 11031 | -0.616 | -0.0213 | No |
| 119 | Selenbp1 | 11085 | -0.633 | -0.0041 | No |
| 120 | Vhl | 11111 | -0.642 | 0.0158 | No |
| 121 | Ndst2 | 11413 | -0.798 | 0.0171 | No |