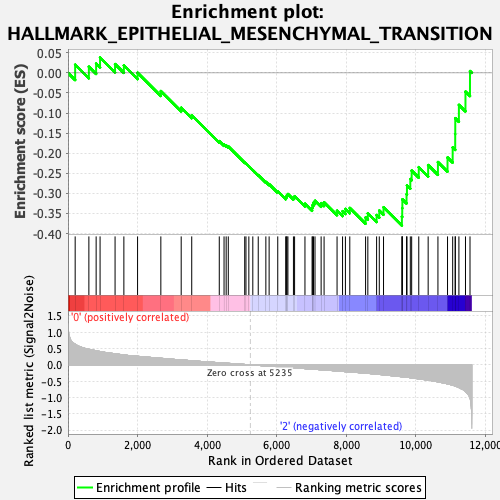
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.3803369 |
| Normalized Enrichment Score (NES) | -1.388599 |
| Nominal p-value | 0.09195402 |
| FDR q-value | 0.44355756 |
| FWER p-Value | 0.607 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pvr | 206 | 0.630 | 0.0204 | No |
| 2 | Tnfaip3 | 600 | 0.476 | 0.0153 | No |
| 3 | Cd44 | 809 | 0.429 | 0.0234 | No |
| 4 | Itgav | 922 | 0.408 | 0.0385 | No |
| 5 | Mcm7 | 1353 | 0.339 | 0.0219 | No |
| 6 | Copa | 1606 | 0.303 | 0.0185 | No |
| 7 | Thbs1 | 2000 | 0.261 | 0.0003 | No |
| 8 | Calu | 2669 | 0.201 | -0.0453 | No |
| 9 | Basp1 | 3254 | 0.150 | -0.0867 | No |
| 10 | Tgfbi | 3557 | 0.124 | -0.1054 | No |
| 11 | Plod1 | 4349 | 0.063 | -0.1700 | No |
| 12 | Tgfb1 | 4486 | 0.054 | -0.1785 | No |
| 13 | Gja1 | 4547 | 0.049 | -0.1807 | No |
| 14 | Jun | 4608 | 0.045 | -0.1831 | No |
| 15 | Sat1 | 5080 | 0.012 | -0.2232 | No |
| 16 | Colgalt1 | 5116 | 0.008 | -0.2257 | No |
| 17 | Mylk | 5199 | 0.003 | -0.2327 | No |
| 18 | Bmp1 | 5313 | -0.005 | -0.2421 | No |
| 19 | Glipr1 | 5469 | -0.016 | -0.2546 | No |
| 20 | Emp3 | 5688 | -0.032 | -0.2715 | No |
| 21 | Pmepa1 | 5782 | -0.039 | -0.2772 | No |
| 22 | Gpc1 | 6030 | -0.056 | -0.2952 | No |
| 23 | Rhob | 6259 | -0.073 | -0.3105 | No |
| 24 | Fuca1 | 6278 | -0.075 | -0.3075 | No |
| 25 | Gadd45b | 6295 | -0.076 | -0.3042 | No |
| 26 | Ecm1 | 6322 | -0.078 | -0.3017 | No |
| 27 | Ntm | 6481 | -0.090 | -0.3100 | No |
| 28 | Vcam1 | 6514 | -0.092 | -0.3072 | No |
| 29 | Ppib | 6812 | -0.115 | -0.3259 | No |
| 30 | Flna | 7017 | -0.129 | -0.3357 | No |
| 31 | Itgb1 | 7033 | -0.130 | -0.3291 | No |
| 32 | Il15 | 7058 | -0.133 | -0.3231 | No |
| 33 | Dst | 7101 | -0.137 | -0.3185 | No |
| 34 | Wipf1 | 7277 | -0.150 | -0.3245 | No |
| 35 | Vegfa | 7362 | -0.156 | -0.3223 | No |
| 36 | Itga2 | 7735 | -0.187 | -0.3431 | No |
| 37 | Sgcb | 7893 | -0.201 | -0.3445 | No |
| 38 | Igfbp4 | 7975 | -0.209 | -0.3389 | No |
| 39 | Lgals1 | 8101 | -0.218 | -0.3364 | No |
| 40 | Itga5 | 8555 | -0.256 | -0.3601 | Yes |
| 41 | P3h1 | 8620 | -0.262 | -0.3497 | Yes |
| 42 | Vegfc | 8872 | -0.286 | -0.3541 | Yes |
| 43 | Plod3 | 8949 | -0.294 | -0.3428 | Yes |
| 44 | Lamc1 | 9071 | -0.305 | -0.3347 | Yes |
| 45 | Tgfbr3 | 9599 | -0.364 | -0.3582 | Yes |
| 46 | Vim | 9609 | -0.364 | -0.3369 | Yes |
| 47 | Pdgfrb | 9613 | -0.365 | -0.3150 | Yes |
| 48 | Notch2 | 9733 | -0.378 | -0.3023 | Yes |
| 49 | Qsox1 | 9745 | -0.380 | -0.2801 | Yes |
| 50 | Ecm2 | 9841 | -0.395 | -0.2643 | Yes |
| 51 | Plod2 | 9880 | -0.400 | -0.2433 | Yes |
| 52 | Itgb3 | 10084 | -0.430 | -0.2348 | Yes |
| 53 | Gadd45a | 10354 | -0.469 | -0.2296 | Yes |
| 54 | Itgb5 | 10635 | -0.522 | -0.2221 | Yes |
| 55 | Plaur | 10911 | -0.580 | -0.2107 | Yes |
| 56 | Tpm1 | 11059 | -0.621 | -0.1857 | Yes |
| 57 | Sntb1 | 11132 | -0.650 | -0.1524 | Yes |
| 58 | Serpine2 | 11134 | -0.651 | -0.1129 | Yes |
| 59 | Tpm4 | 11238 | -0.694 | -0.0797 | Yes |
| 60 | Fas | 11427 | -0.812 | -0.0466 | Yes |
| 61 | Capg | 11557 | -1.025 | 0.0045 | Yes |

