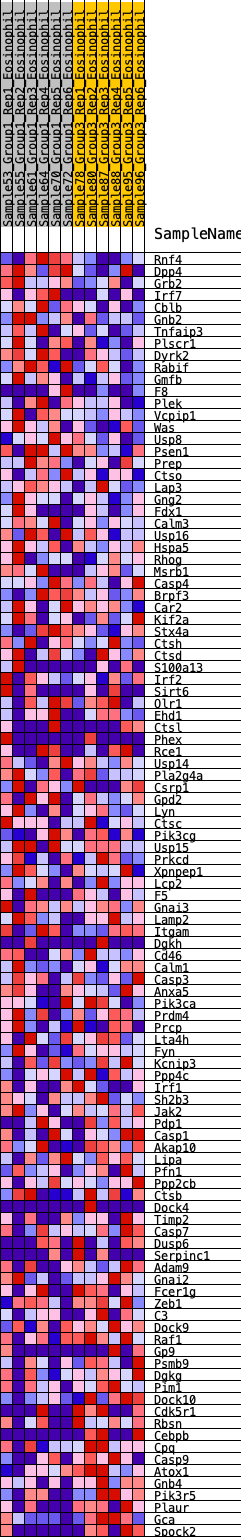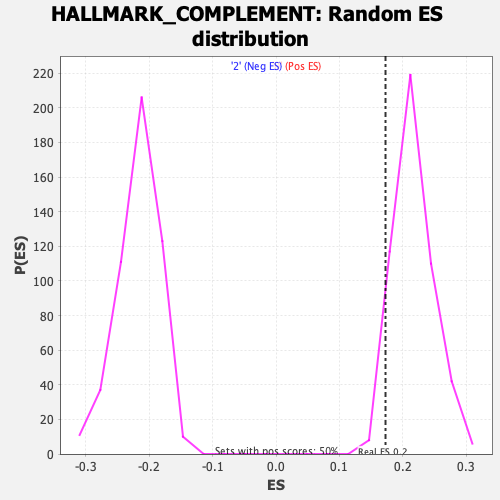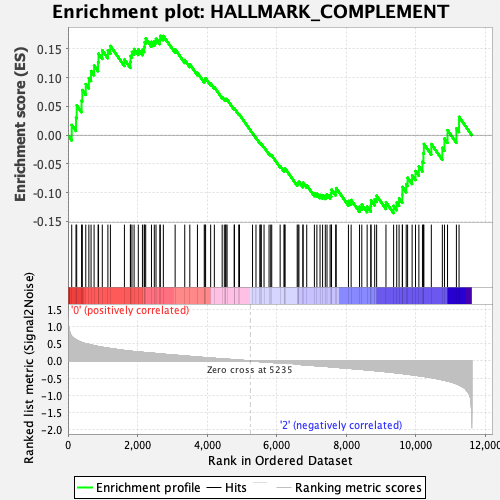
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | 0.17274047 |
| Normalized Enrichment Score (NES) | 0.7969647 |
| Nominal p-value | 0.9482072 |
| FDR q-value | 0.8912996 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rnf4 | 107 | 0.714 | 0.0179 | Yes |
| 2 | Dpp4 | 230 | 0.615 | 0.0307 | Yes |
| 3 | Grb2 | 250 | 0.602 | 0.0519 | Yes |
| 4 | Irf7 | 390 | 0.536 | 0.0603 | Yes |
| 5 | Cblb | 412 | 0.529 | 0.0786 | Yes |
| 6 | Gnb2 | 511 | 0.497 | 0.0890 | Yes |
| 7 | Tnfaip3 | 600 | 0.476 | 0.0994 | Yes |
| 8 | Plscr1 | 663 | 0.463 | 0.1117 | Yes |
| 9 | Dyrk2 | 750 | 0.443 | 0.1211 | Yes |
| 10 | Rabif | 865 | 0.418 | 0.1271 | Yes |
| 11 | Gmfb | 878 | 0.415 | 0.1418 | Yes |
| 12 | F8 | 986 | 0.396 | 0.1476 | Yes |
| 13 | Plek | 1147 | 0.369 | 0.1477 | Yes |
| 14 | Vcpip1 | 1219 | 0.358 | 0.1552 | Yes |
| 15 | Was | 1622 | 0.301 | 0.1317 | Yes |
| 16 | Usp8 | 1793 | 0.280 | 0.1276 | Yes |
| 17 | Psen1 | 1799 | 0.280 | 0.1378 | Yes |
| 18 | Prep | 1839 | 0.276 | 0.1449 | Yes |
| 19 | Ctso | 1900 | 0.270 | 0.1500 | Yes |
| 20 | Lap3 | 2021 | 0.259 | 0.1494 | Yes |
| 21 | Gng2 | 2140 | 0.248 | 0.1486 | Yes |
| 22 | Fdx1 | 2190 | 0.243 | 0.1536 | Yes |
| 23 | Calm3 | 2204 | 0.242 | 0.1617 | Yes |
| 24 | Usp16 | 2234 | 0.239 | 0.1682 | Yes |
| 25 | Hspa5 | 2400 | 0.224 | 0.1624 | Yes |
| 26 | Rhog | 2481 | 0.218 | 0.1638 | Yes |
| 27 | Msrb1 | 2530 | 0.214 | 0.1678 | Yes |
| 28 | Casp4 | 2641 | 0.204 | 0.1660 | Yes |
| 29 | Brpf3 | 2653 | 0.203 | 0.1727 | Yes |
| 30 | Car2 | 2741 | 0.195 | 0.1726 | No |
| 31 | Kif2a | 3080 | 0.164 | 0.1494 | No |
| 32 | Stx4a | 3356 | 0.141 | 0.1309 | No |
| 33 | Ctsh | 3502 | 0.130 | 0.1232 | No |
| 34 | Ctsd | 3721 | 0.111 | 0.1085 | No |
| 35 | S100a13 | 3917 | 0.097 | 0.0953 | No |
| 36 | Irf2 | 3937 | 0.096 | 0.0972 | No |
| 37 | Sirt6 | 3956 | 0.094 | 0.0993 | No |
| 38 | Olr1 | 4101 | 0.083 | 0.0899 | No |
| 39 | Ehd1 | 4207 | 0.075 | 0.0836 | No |
| 40 | Ctsl | 4432 | 0.057 | 0.0663 | No |
| 41 | Phex | 4494 | 0.053 | 0.0630 | No |
| 42 | Rce1 | 4534 | 0.050 | 0.0616 | No |
| 43 | Usp14 | 4539 | 0.050 | 0.0631 | No |
| 44 | Pla2g4a | 4573 | 0.048 | 0.0621 | No |
| 45 | Csrp1 | 4777 | 0.035 | 0.0457 | No |
| 46 | Gpd2 | 4786 | 0.034 | 0.0463 | No |
| 47 | Lyn | 4909 | 0.024 | 0.0366 | No |
| 48 | Ctsc | 4930 | 0.022 | 0.0357 | No |
| 49 | Pik3cg | 5305 | -0.005 | 0.0034 | No |
| 50 | Usp15 | 5404 | -0.012 | -0.0047 | No |
| 51 | Prkcd | 5515 | -0.020 | -0.0135 | No |
| 52 | Xpnpep1 | 5540 | -0.022 | -0.0147 | No |
| 53 | Lcp2 | 5557 | -0.023 | -0.0152 | No |
| 54 | F5 | 5636 | -0.029 | -0.0209 | No |
| 55 | Gnai3 | 5780 | -0.039 | -0.0319 | No |
| 56 | Lamp2 | 5828 | -0.042 | -0.0344 | No |
| 57 | Itgam | 5857 | -0.044 | -0.0351 | No |
| 58 | Dgkh | 6099 | -0.061 | -0.0538 | No |
| 59 | Cd46 | 6211 | -0.070 | -0.0608 | No |
| 60 | Calm1 | 6219 | -0.070 | -0.0587 | No |
| 61 | Casp3 | 6242 | -0.072 | -0.0579 | No |
| 62 | Anxa5 | 6589 | -0.097 | -0.0843 | No |
| 63 | Pik3ca | 6615 | -0.099 | -0.0827 | No |
| 64 | Prdm4 | 6639 | -0.101 | -0.0808 | No |
| 65 | Prcp | 6753 | -0.110 | -0.0864 | No |
| 66 | Lta4h | 6758 | -0.111 | -0.0826 | No |
| 67 | Fyn | 6863 | -0.120 | -0.0871 | No |
| 68 | Kcnip3 | 7083 | -0.135 | -0.1010 | No |
| 69 | Ppp4c | 7155 | -0.140 | -0.1018 | No |
| 70 | Irf1 | 7246 | -0.148 | -0.1040 | No |
| 71 | Sh2b3 | 7319 | -0.153 | -0.1044 | No |
| 72 | Jak2 | 7397 | -0.159 | -0.1051 | No |
| 73 | Pdp1 | 7446 | -0.161 | -0.1031 | No |
| 74 | Casp1 | 7535 | -0.170 | -0.1043 | No |
| 75 | Akap10 | 7569 | -0.173 | -0.1006 | No |
| 76 | Lipa | 7576 | -0.173 | -0.0945 | No |
| 77 | Pfn1 | 7696 | -0.183 | -0.0979 | No |
| 78 | Ppp2cb | 7710 | -0.185 | -0.0920 | No |
| 79 | Ctsb | 8062 | -0.214 | -0.1143 | No |
| 80 | Dock4 | 8140 | -0.221 | -0.1126 | No |
| 81 | Timp2 | 8381 | -0.240 | -0.1244 | No |
| 82 | Casp7 | 8446 | -0.246 | -0.1206 | No |
| 83 | Dusp6 | 8600 | -0.260 | -0.1240 | No |
| 84 | Serpinc1 | 8706 | -0.269 | -0.1229 | No |
| 85 | Adam9 | 8711 | -0.269 | -0.1130 | No |
| 86 | Gnai2 | 8817 | -0.281 | -0.1114 | No |
| 87 | Fcer1g | 8873 | -0.287 | -0.1053 | No |
| 88 | Zeb1 | 9141 | -0.313 | -0.1166 | No |
| 89 | C3 | 9361 | -0.336 | -0.1228 | No |
| 90 | Dock9 | 9450 | -0.347 | -0.1173 | No |
| 91 | Raf1 | 9518 | -0.354 | -0.1096 | No |
| 92 | Gp9 | 9612 | -0.365 | -0.1038 | No |
| 93 | Psmb9 | 9614 | -0.365 | -0.0900 | No |
| 94 | Dgkg | 9723 | -0.378 | -0.0850 | No |
| 95 | Pim1 | 9763 | -0.382 | -0.0739 | No |
| 96 | Dock10 | 9893 | -0.402 | -0.0698 | No |
| 97 | Cdk5r1 | 9990 | -0.417 | -0.0623 | No |
| 98 | Rbsn | 10087 | -0.430 | -0.0543 | No |
| 99 | Cebpb | 10190 | -0.447 | -0.0461 | No |
| 100 | Cpq | 10218 | -0.450 | -0.0314 | No |
| 101 | Casp9 | 10231 | -0.451 | -0.0152 | No |
| 102 | Atox1 | 10445 | -0.484 | -0.0153 | No |
| 103 | Gnb4 | 10762 | -0.548 | -0.0220 | No |
| 104 | Pik3r5 | 10823 | -0.562 | -0.0058 | No |
| 105 | Plaur | 10911 | -0.580 | 0.0087 | No |
| 106 | Gca | 11166 | -0.663 | 0.0119 | No |
| 107 | Spock2 | 11242 | -0.698 | 0.0319 | No |