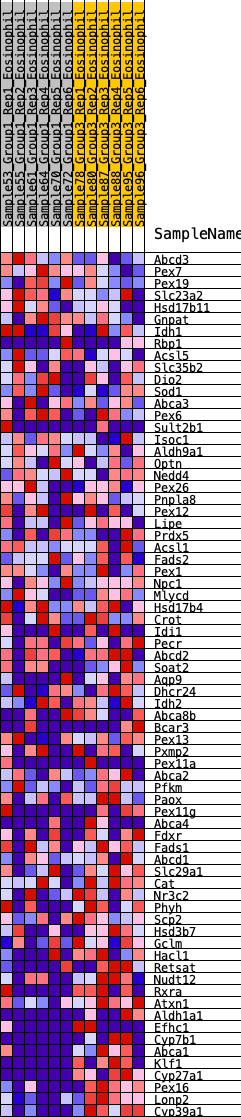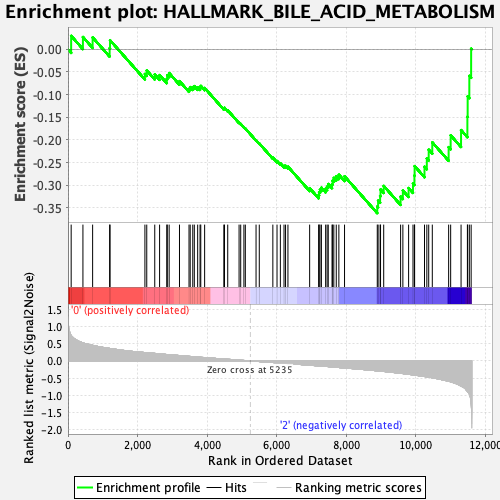
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.3619515 |
| Normalized Enrichment Score (NES) | -1.3472998 |
| Nominal p-value | 0.10982659 |
| FDR q-value | 0.2674823 |
| FWER p-Value | 0.703 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcd3 | 91 | 0.736 | 0.0299 | No |
| 2 | Pex7 | 429 | 0.521 | 0.0273 | No |
| 3 | Pex19 | 708 | 0.452 | 0.0264 | No |
| 4 | Slc23a2 | 1197 | 0.361 | 0.0026 | No |
| 5 | Hsd17b11 | 1209 | 0.360 | 0.0201 | No |
| 6 | Gnpat | 2211 | 0.241 | -0.0543 | No |
| 7 | Idh1 | 2264 | 0.237 | -0.0467 | No |
| 8 | Rbp1 | 2494 | 0.217 | -0.0554 | No |
| 9 | Acsl5 | 2633 | 0.205 | -0.0569 | No |
| 10 | Slc35b2 | 2840 | 0.184 | -0.0653 | No |
| 11 | Dio2 | 2854 | 0.183 | -0.0570 | No |
| 12 | Sod1 | 2908 | 0.179 | -0.0525 | No |
| 13 | Abca3 | 3205 | 0.154 | -0.0702 | No |
| 14 | Pex6 | 3479 | 0.131 | -0.0872 | No |
| 15 | Sult2b1 | 3513 | 0.129 | -0.0834 | No |
| 16 | Isoc1 | 3587 | 0.121 | -0.0835 | No |
| 17 | Aldh9a1 | 3631 | 0.118 | -0.0812 | No |
| 18 | Optn | 3728 | 0.111 | -0.0839 | No |
| 19 | Nedd4 | 3796 | 0.106 | -0.0843 | No |
| 20 | Pex26 | 3817 | 0.104 | -0.0807 | No |
| 21 | Pnpla8 | 3928 | 0.096 | -0.0853 | No |
| 22 | Pex12 | 4480 | 0.054 | -0.1303 | No |
| 23 | Lipe | 4499 | 0.053 | -0.1291 | No |
| 24 | Prdx5 | 4592 | 0.046 | -0.1347 | No |
| 25 | Acsl1 | 4918 | 0.023 | -0.1617 | No |
| 26 | Fads2 | 4965 | 0.019 | -0.1647 | No |
| 27 | Pex1 | 5057 | 0.013 | -0.1720 | No |
| 28 | Npc1 | 5098 | 0.010 | -0.1749 | No |
| 29 | Mlycd | 5406 | -0.012 | -0.2009 | No |
| 30 | Hsd17b4 | 5499 | -0.019 | -0.2080 | No |
| 31 | Crot | 5889 | -0.046 | -0.2393 | No |
| 32 | Idi1 | 6009 | -0.055 | -0.2468 | No |
| 33 | Pecr | 6105 | -0.061 | -0.2519 | No |
| 34 | Abcd2 | 6210 | -0.070 | -0.2574 | No |
| 35 | Soat2 | 6254 | -0.073 | -0.2573 | No |
| 36 | Aqp9 | 6323 | -0.078 | -0.2592 | No |
| 37 | Dhcr24 | 6947 | -0.124 | -0.3069 | No |
| 38 | Idh2 | 7206 | -0.145 | -0.3218 | No |
| 39 | Abca8b | 7219 | -0.146 | -0.3154 | No |
| 40 | Bcar3 | 7244 | -0.148 | -0.3099 | No |
| 41 | Pex13 | 7285 | -0.151 | -0.3056 | No |
| 42 | Pxmp2 | 7407 | -0.159 | -0.3079 | No |
| 43 | Pex11a | 7449 | -0.162 | -0.3032 | No |
| 44 | Abca2 | 7482 | -0.164 | -0.2975 | No |
| 45 | Pfkm | 7591 | -0.175 | -0.2979 | No |
| 46 | Paox | 7607 | -0.176 | -0.2902 | No |
| 47 | Pex11g | 7642 | -0.179 | -0.2839 | No |
| 48 | Abca4 | 7712 | -0.185 | -0.2804 | No |
| 49 | Fdxr | 7787 | -0.192 | -0.2770 | No |
| 50 | Fads1 | 7950 | -0.206 | -0.2805 | No |
| 51 | Abcd1 | 8891 | -0.288 | -0.3472 | Yes |
| 52 | Slc29a1 | 8915 | -0.292 | -0.3342 | Yes |
| 53 | Cat | 8973 | -0.296 | -0.3240 | Yes |
| 54 | Nr3c2 | 8985 | -0.298 | -0.3097 | Yes |
| 55 | Phyh | 9076 | -0.306 | -0.3018 | Yes |
| 56 | Scp2 | 9562 | -0.361 | -0.3254 | Yes |
| 57 | Hsd3b7 | 9627 | -0.367 | -0.3121 | Yes |
| 58 | Gclm | 9792 | -0.387 | -0.3065 | Yes |
| 59 | Hacl1 | 9918 | -0.407 | -0.2964 | Yes |
| 60 | Retsat | 9959 | -0.412 | -0.2788 | Yes |
| 61 | Nudt12 | 9965 | -0.413 | -0.2581 | Yes |
| 62 | Rxra | 10249 | -0.453 | -0.2594 | Yes |
| 63 | Atxn1 | 10316 | -0.465 | -0.2413 | Yes |
| 64 | Aldh1a1 | 10370 | -0.471 | -0.2217 | Yes |
| 65 | Efhc1 | 10473 | -0.488 | -0.2055 | Yes |
| 66 | Cyp7b1 | 10943 | -0.587 | -0.2161 | Yes |
| 67 | Abca1 | 11001 | -0.606 | -0.1900 | Yes |
| 68 | Klf1 | 11300 | -0.730 | -0.1784 | Yes |
| 69 | Cyp27a1 | 11482 | -0.882 | -0.1489 | Yes |
| 70 | Pex16 | 11490 | -0.891 | -0.1038 | Yes |
| 71 | Lonp2 | 11538 | -0.963 | -0.0585 | Yes |
| 72 | Cyp39a1 | 11590 | -1.260 | 0.0016 | Yes |