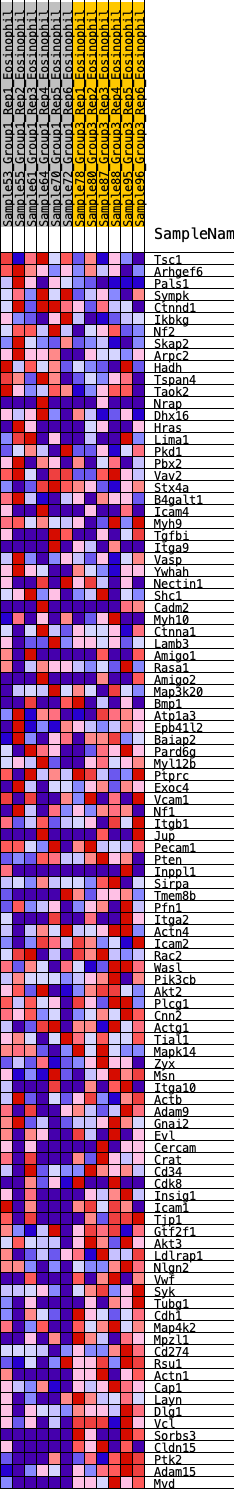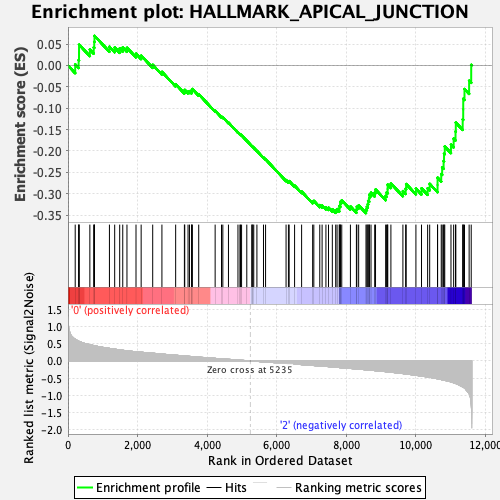
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group3.Eosinophil_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | -0.34539014 |
| Normalized Enrichment Score (NES) | -1.3645482 |
| Nominal p-value | 0.072874494 |
| FDR q-value | 0.35824734 |
| FWER p-Value | 0.659 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tsc1 | 207 | 0.628 | 0.0025 | No |
| 2 | Arhgef6 | 306 | 0.571 | 0.0126 | No |
| 3 | Pals1 | 317 | 0.567 | 0.0303 | No |
| 4 | Sympk | 318 | 0.567 | 0.0487 | No |
| 5 | Ctnnd1 | 628 | 0.470 | 0.0372 | No |
| 6 | Ikbkg | 739 | 0.445 | 0.0422 | No |
| 7 | Nf2 | 755 | 0.443 | 0.0553 | No |
| 8 | Skap2 | 762 | 0.442 | 0.0692 | No |
| 9 | Arpc2 | 1191 | 0.362 | 0.0439 | No |
| 10 | Hadh | 1342 | 0.341 | 0.0419 | No |
| 11 | Tspan4 | 1486 | 0.319 | 0.0399 | No |
| 12 | Taok2 | 1574 | 0.308 | 0.0424 | No |
| 13 | Nrap | 1695 | 0.291 | 0.0415 | No |
| 14 | Dhx16 | 1955 | 0.265 | 0.0276 | No |
| 15 | Hras | 2103 | 0.252 | 0.0231 | No |
| 16 | Lima1 | 2435 | 0.222 | 0.0015 | No |
| 17 | Pkd1 | 2700 | 0.198 | -0.0149 | No |
| 18 | Pbx2 | 3095 | 0.163 | -0.0439 | No |
| 19 | Vav2 | 3346 | 0.142 | -0.0610 | No |
| 20 | Stx4a | 3356 | 0.141 | -0.0571 | No |
| 21 | B4galt1 | 3451 | 0.133 | -0.0609 | No |
| 22 | Icam4 | 3492 | 0.130 | -0.0602 | No |
| 23 | Myh9 | 3553 | 0.125 | -0.0613 | No |
| 24 | Tgfbi | 3557 | 0.124 | -0.0575 | No |
| 25 | Itga9 | 3575 | 0.122 | -0.0550 | No |
| 26 | Vasp | 3758 | 0.108 | -0.0673 | No |
| 27 | Ywhah | 4230 | 0.072 | -0.1058 | No |
| 28 | Nectin1 | 4413 | 0.059 | -0.1197 | No |
| 29 | Shc1 | 4451 | 0.056 | -0.1211 | No |
| 30 | Cadm2 | 4613 | 0.045 | -0.1337 | No |
| 31 | Myh10 | 4886 | 0.026 | -0.1564 | No |
| 32 | Ctnna1 | 4943 | 0.021 | -0.1606 | No |
| 33 | Lamb3 | 4959 | 0.019 | -0.1613 | No |
| 34 | Amigo1 | 4990 | 0.017 | -0.1634 | No |
| 35 | Rasa1 | 5140 | 0.006 | -0.1761 | No |
| 36 | Amigo2 | 5286 | -0.003 | -0.1886 | No |
| 37 | Map3k20 | 5292 | -0.004 | -0.1889 | No |
| 38 | Bmp1 | 5313 | -0.005 | -0.1905 | No |
| 39 | Atp1a3 | 5340 | -0.007 | -0.1925 | No |
| 40 | Epb41l2 | 5431 | -0.013 | -0.1999 | No |
| 41 | Baiap2 | 5620 | -0.027 | -0.2153 | No |
| 42 | Pard6g | 5680 | -0.031 | -0.2194 | No |
| 43 | Myl12b | 6269 | -0.074 | -0.2681 | No |
| 44 | Ptprc | 6337 | -0.079 | -0.2714 | No |
| 45 | Exoc4 | 6358 | -0.081 | -0.2705 | No |
| 46 | Vcam1 | 6514 | -0.092 | -0.2810 | No |
| 47 | Nf1 | 6716 | -0.107 | -0.2949 | No |
| 48 | Itgb1 | 7033 | -0.130 | -0.3182 | No |
| 49 | Jup | 7067 | -0.134 | -0.3167 | No |
| 50 | Pecam1 | 7238 | -0.147 | -0.3266 | No |
| 51 | Pten | 7305 | -0.152 | -0.3274 | No |
| 52 | Inppl1 | 7415 | -0.160 | -0.3317 | No |
| 53 | Sirpa | 7489 | -0.165 | -0.3326 | No |
| 54 | Tmem8b | 7599 | -0.176 | -0.3363 | No |
| 55 | Pfn1 | 7696 | -0.183 | -0.3387 | Yes |
| 56 | Itga2 | 7735 | -0.187 | -0.3359 | Yes |
| 57 | Actn4 | 7800 | -0.193 | -0.3352 | Yes |
| 58 | Icam2 | 7801 | -0.193 | -0.3289 | Yes |
| 59 | Rac2 | 7828 | -0.195 | -0.3248 | Yes |
| 60 | Wasl | 7832 | -0.195 | -0.3186 | Yes |
| 61 | Pik3cb | 7870 | -0.199 | -0.3154 | Yes |
| 62 | Akt2 | 8119 | -0.219 | -0.3298 | Yes |
| 63 | Plcg1 | 8291 | -0.232 | -0.3370 | Yes |
| 64 | Cnn2 | 8304 | -0.233 | -0.3305 | Yes |
| 65 | Actg1 | 8356 | -0.238 | -0.3271 | Yes |
| 66 | Tial1 | 8567 | -0.257 | -0.3370 | Yes |
| 67 | Mapk14 | 8592 | -0.259 | -0.3306 | Yes |
| 68 | Zyx | 8619 | -0.262 | -0.3243 | Yes |
| 69 | Msn | 8630 | -0.262 | -0.3166 | Yes |
| 70 | Itga10 | 8659 | -0.265 | -0.3104 | Yes |
| 71 | Actb | 8663 | -0.265 | -0.3020 | Yes |
| 72 | Adam9 | 8711 | -0.269 | -0.2973 | Yes |
| 73 | Gnai2 | 8817 | -0.281 | -0.2973 | Yes |
| 74 | Evl | 8841 | -0.283 | -0.2901 | Yes |
| 75 | Cercam | 9132 | -0.312 | -0.3051 | Yes |
| 76 | Crat | 9159 | -0.315 | -0.2970 | Yes |
| 77 | Cd34 | 9189 | -0.319 | -0.2892 | Yes |
| 78 | Cdk8 | 9190 | -0.319 | -0.2787 | Yes |
| 79 | Insig1 | 9283 | -0.330 | -0.2760 | Yes |
| 80 | Icam1 | 9628 | -0.367 | -0.2939 | Yes |
| 81 | Tjp1 | 9707 | -0.376 | -0.2884 | Yes |
| 82 | Gtf2f1 | 9722 | -0.377 | -0.2773 | Yes |
| 83 | Akt3 | 10002 | -0.418 | -0.2879 | Yes |
| 84 | Ldlrap1 | 10163 | -0.442 | -0.2874 | Yes |
| 85 | Nlgn2 | 10339 | -0.467 | -0.2873 | Yes |
| 86 | Vwf | 10398 | -0.475 | -0.2769 | Yes |
| 87 | Syk | 10624 | -0.520 | -0.2794 | Yes |
| 88 | Tubg1 | 10625 | -0.521 | -0.2625 | Yes |
| 89 | Cdh1 | 10730 | -0.541 | -0.2538 | Yes |
| 90 | Map4k2 | 10757 | -0.547 | -0.2382 | Yes |
| 91 | Mpzl1 | 10798 | -0.557 | -0.2235 | Yes |
| 92 | Cd274 | 10811 | -0.560 | -0.2063 | Yes |
| 93 | Rsu1 | 10831 | -0.564 | -0.1895 | Yes |
| 94 | Actn1 | 11010 | -0.608 | -0.1852 | Yes |
| 95 | Cap1 | 11087 | -0.633 | -0.1711 | Yes |
| 96 | Layn | 11141 | -0.653 | -0.1544 | Yes |
| 97 | Dlg1 | 11149 | -0.656 | -0.1336 | Yes |
| 98 | Vcl | 11347 | -0.752 | -0.1262 | Yes |
| 99 | Sorbs3 | 11362 | -0.761 | -0.1026 | Yes |
| 100 | Cldn15 | 11363 | -0.761 | -0.0777 | Yes |
| 101 | Ptk2 | 11399 | -0.786 | -0.0551 | Yes |
| 102 | Adam15 | 11532 | -0.950 | -0.0355 | Yes |
| 103 | Mvd | 11592 | -1.291 | 0.0015 | Yes |