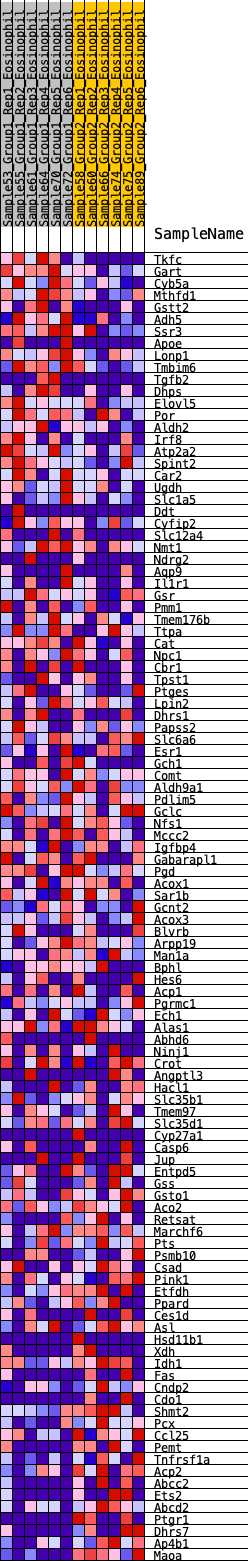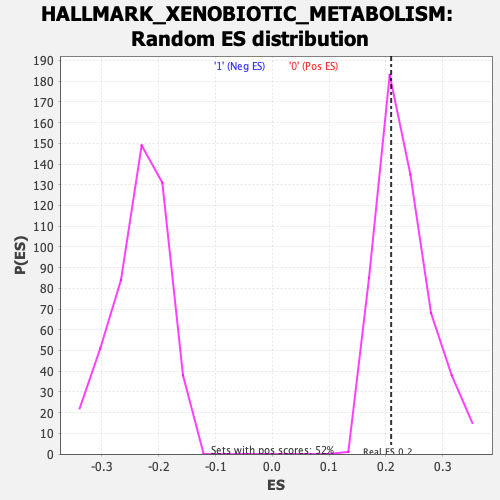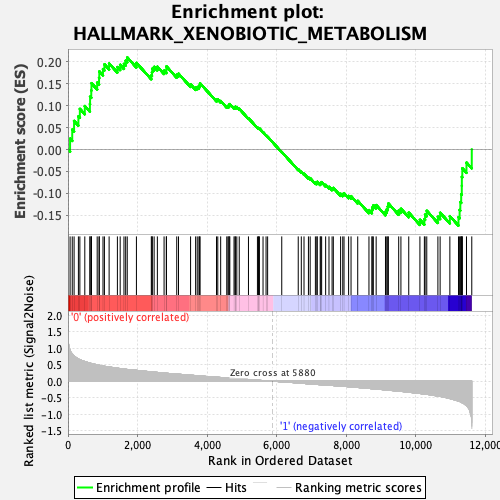
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | 0.20939927 |
| Normalized Enrichment Score (NES) | 0.90089124 |
| Nominal p-value | 0.6514286 |
| FDR q-value | 0.8335669 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tkfc | 60 | 0.949 | 0.0247 | Yes |
| 2 | Gart | 122 | 0.826 | 0.0454 | Yes |
| 3 | Cyb5a | 175 | 0.765 | 0.0650 | Yes |
| 4 | Mthfd1 | 299 | 0.673 | 0.0755 | Yes |
| 5 | Gstt2 | 341 | 0.648 | 0.0923 | Yes |
| 6 | Adh5 | 483 | 0.595 | 0.0988 | Yes |
| 7 | Ssr3 | 630 | 0.548 | 0.1034 | Yes |
| 8 | Apoe | 635 | 0.547 | 0.1203 | Yes |
| 9 | Lonp1 | 669 | 0.537 | 0.1343 | Yes |
| 10 | Tmbim6 | 675 | 0.534 | 0.1507 | Yes |
| 11 | Tgfb2 | 839 | 0.498 | 0.1522 | Yes |
| 12 | Dhps | 893 | 0.484 | 0.1629 | Yes |
| 13 | Elovl5 | 900 | 0.482 | 0.1775 | Yes |
| 14 | Por | 1006 | 0.460 | 0.1829 | Yes |
| 15 | Aldh2 | 1047 | 0.452 | 0.1937 | Yes |
| 16 | Irf8 | 1182 | 0.431 | 0.1956 | Yes |
| 17 | Atp2a2 | 1420 | 0.395 | 0.1874 | Yes |
| 18 | Spint2 | 1500 | 0.382 | 0.1926 | Yes |
| 19 | Car2 | 1609 | 0.368 | 0.1948 | Yes |
| 20 | Ugdh | 1658 | 0.361 | 0.2020 | Yes |
| 21 | Slc1a5 | 1703 | 0.356 | 0.2094 | Yes |
| 22 | Ddt | 1967 | 0.329 | 0.1969 | No |
| 23 | Cyfip2 | 2395 | 0.281 | 0.1686 | No |
| 24 | Slc12a4 | 2414 | 0.279 | 0.1758 | No |
| 25 | Nmt1 | 2423 | 0.278 | 0.1839 | No |
| 26 | Ndrg2 | 2476 | 0.273 | 0.1880 | No |
| 27 | Aqp9 | 2569 | 0.266 | 0.1883 | No |
| 28 | Il1r1 | 2762 | 0.247 | 0.1794 | No |
| 29 | Gsr | 2828 | 0.242 | 0.1814 | No |
| 30 | Pmm1 | 2829 | 0.242 | 0.1890 | No |
| 31 | Tmem176b | 3126 | 0.216 | 0.1700 | No |
| 32 | Ttpa | 3177 | 0.211 | 0.1723 | No |
| 33 | Cat | 3524 | 0.182 | 0.1480 | No |
| 34 | Npc1 | 3669 | 0.170 | 0.1408 | No |
| 35 | Cbr1 | 3715 | 0.167 | 0.1422 | No |
| 36 | Tpst1 | 3764 | 0.162 | 0.1431 | No |
| 37 | Ptges | 3784 | 0.160 | 0.1465 | No |
| 38 | Lpin2 | 3796 | 0.159 | 0.1505 | No |
| 39 | Dhrs1 | 4269 | 0.121 | 0.1133 | No |
| 40 | Papss2 | 4306 | 0.118 | 0.1139 | No |
| 41 | Slc6a6 | 4389 | 0.113 | 0.1103 | No |
| 42 | Esr1 | 4564 | 0.101 | 0.0984 | No |
| 43 | Gch1 | 4598 | 0.098 | 0.0986 | No |
| 44 | Comt | 4628 | 0.096 | 0.0991 | No |
| 45 | Aldh9a1 | 4629 | 0.096 | 0.1021 | No |
| 46 | Pdlim5 | 4652 | 0.094 | 0.1032 | No |
| 47 | Gclc | 4773 | 0.085 | 0.0954 | No |
| 48 | Nfs1 | 4803 | 0.081 | 0.0955 | No |
| 49 | Mccc2 | 4812 | 0.081 | 0.0973 | No |
| 50 | Igfbp4 | 4853 | 0.077 | 0.0963 | No |
| 51 | Gabarapl1 | 4923 | 0.072 | 0.0925 | No |
| 52 | Pgd | 5188 | 0.051 | 0.0712 | No |
| 53 | Acox1 | 5446 | 0.031 | 0.0498 | No |
| 54 | Sar1b | 5479 | 0.029 | 0.0479 | No |
| 55 | Gcnt2 | 5482 | 0.029 | 0.0487 | No |
| 56 | Acox3 | 5503 | 0.027 | 0.0478 | No |
| 57 | Blvrb | 5607 | 0.019 | 0.0394 | No |
| 58 | Arpp19 | 5694 | 0.014 | 0.0324 | No |
| 59 | Man1a | 5739 | 0.011 | 0.0289 | No |
| 60 | Bphl | 6145 | -0.020 | -0.0057 | No |
| 61 | Hes6 | 6619 | -0.054 | -0.0451 | No |
| 62 | Acp1 | 6707 | -0.062 | -0.0507 | No |
| 63 | Pgrmc1 | 6785 | -0.068 | -0.0553 | No |
| 64 | Ech1 | 6912 | -0.079 | -0.0637 | No |
| 65 | Alas1 | 6962 | -0.083 | -0.0654 | No |
| 66 | Abhd6 | 7112 | -0.096 | -0.0753 | No |
| 67 | Ninj1 | 7154 | -0.098 | -0.0758 | No |
| 68 | Crot | 7165 | -0.099 | -0.0735 | No |
| 69 | Angptl3 | 7250 | -0.104 | -0.0775 | No |
| 70 | Hacl1 | 7268 | -0.105 | -0.0757 | No |
| 71 | Slc35b1 | 7292 | -0.107 | -0.0743 | No |
| 72 | Tmem97 | 7407 | -0.117 | -0.0805 | No |
| 73 | Slc35d1 | 7503 | -0.122 | -0.0849 | No |
| 74 | Cyp27a1 | 7592 | -0.130 | -0.0885 | No |
| 75 | Casp6 | 7628 | -0.131 | -0.0874 | No |
| 76 | Jup | 7837 | -0.148 | -0.1008 | No |
| 77 | Entpd5 | 7899 | -0.152 | -0.1013 | No |
| 78 | Gss | 7935 | -0.156 | -0.0995 | No |
| 79 | Gsto1 | 8067 | -0.166 | -0.1056 | No |
| 80 | Aco2 | 8137 | -0.172 | -0.1062 | No |
| 81 | Retsat | 8329 | -0.190 | -0.1168 | No |
| 82 | Marchf6 | 8652 | -0.219 | -0.1379 | No |
| 83 | Pts | 8737 | -0.227 | -0.1381 | No |
| 84 | Psmb10 | 8747 | -0.228 | -0.1317 | No |
| 85 | Csad | 8778 | -0.231 | -0.1270 | No |
| 86 | Pink1 | 8859 | -0.238 | -0.1265 | No |
| 87 | Etfdh | 9127 | -0.265 | -0.1414 | No |
| 88 | Ppard | 9164 | -0.269 | -0.1360 | No |
| 89 | Ces1d | 9192 | -0.271 | -0.1298 | No |
| 90 | Asl | 9210 | -0.273 | -0.1227 | No |
| 91 | Hsd11b1 | 9507 | -0.301 | -0.1390 | No |
| 92 | Xdh | 9571 | -0.308 | -0.1348 | No |
| 93 | Idh1 | 9796 | -0.334 | -0.1437 | No |
| 94 | Fas | 10117 | -0.371 | -0.1598 | No |
| 95 | Cndp2 | 10245 | -0.391 | -0.1586 | No |
| 96 | Cdo1 | 10266 | -0.393 | -0.1479 | No |
| 97 | Shmt2 | 10314 | -0.400 | -0.1394 | No |
| 98 | Pcx | 10634 | -0.450 | -0.1529 | No |
| 99 | Ccl25 | 10698 | -0.460 | -0.1439 | No |
| 100 | Pemt | 10978 | -0.526 | -0.1516 | No |
| 101 | Tnfrsf1a | 11226 | -0.606 | -0.1540 | No |
| 102 | Acp2 | 11259 | -0.614 | -0.1374 | No |
| 103 | Abcc2 | 11279 | -0.621 | -0.1195 | No |
| 104 | Ets2 | 11304 | -0.634 | -0.1016 | No |
| 105 | Abcd2 | 11321 | -0.646 | -0.0827 | No |
| 106 | Ptgr1 | 11323 | -0.649 | -0.0623 | No |
| 107 | Dhrs7 | 11336 | -0.653 | -0.0428 | No |
| 108 | Ap4b1 | 11456 | -0.738 | -0.0299 | No |
| 109 | Maoa | 11608 | -1.367 | 0.0001 | No |