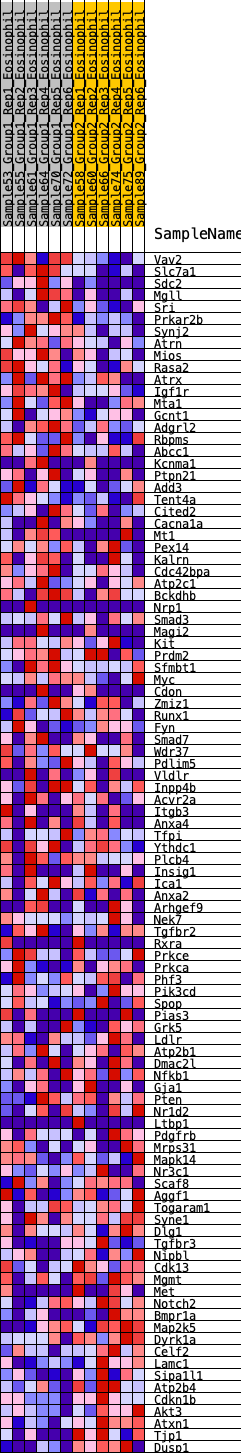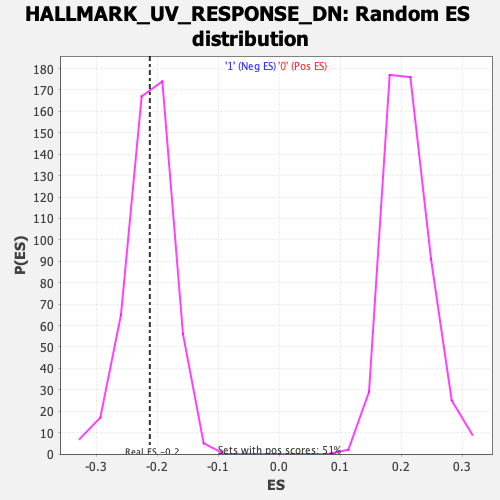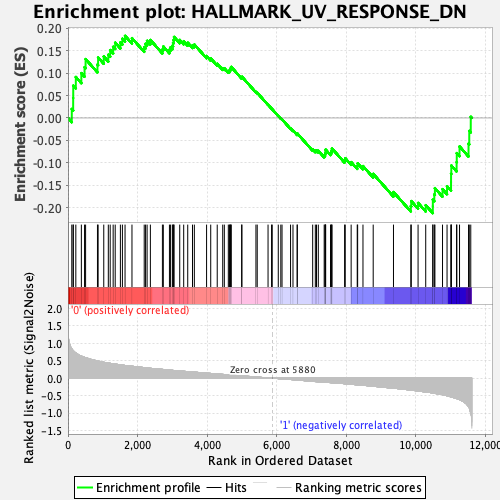
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | -0.2119561 |
| Normalized Enrichment Score (NES) | -0.9923103 |
| Nominal p-value | 0.48676172 |
| FDR q-value | 0.61118186 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vav2 | 109 | 0.841 | 0.0202 | No |
| 2 | Slc7a1 | 149 | 0.788 | 0.0445 | No |
| 3 | Sdc2 | 151 | 0.786 | 0.0721 | No |
| 4 | Mgll | 225 | 0.725 | 0.0913 | No |
| 5 | Sri | 384 | 0.635 | 0.0999 | No |
| 6 | Prkar2b | 476 | 0.598 | 0.1131 | No |
| 7 | Synj2 | 506 | 0.585 | 0.1312 | No |
| 8 | Atrn | 847 | 0.496 | 0.1191 | No |
| 9 | Mios | 866 | 0.491 | 0.1348 | No |
| 10 | Rasa2 | 1028 | 0.456 | 0.1369 | No |
| 11 | Atrx | 1159 | 0.436 | 0.1409 | No |
| 12 | Igf1r | 1213 | 0.425 | 0.1513 | No |
| 13 | Mta1 | 1298 | 0.413 | 0.1586 | No |
| 14 | Gcnt1 | 1357 | 0.406 | 0.1678 | No |
| 15 | Adgrl2 | 1506 | 0.381 | 0.1684 | No |
| 16 | Rbpms | 1564 | 0.374 | 0.1766 | No |
| 17 | Abcc1 | 1640 | 0.364 | 0.1829 | No |
| 18 | Kcnma1 | 1839 | 0.342 | 0.1778 | No |
| 19 | Ptpn21 | 2191 | 0.303 | 0.1579 | No |
| 20 | Add3 | 2230 | 0.300 | 0.1652 | No |
| 21 | Tent4a | 2274 | 0.295 | 0.1719 | No |
| 22 | Cited2 | 2368 | 0.283 | 0.1738 | No |
| 23 | Cacna1a | 2714 | 0.252 | 0.1527 | No |
| 24 | Mt1 | 2740 | 0.249 | 0.1593 | No |
| 25 | Pex14 | 2916 | 0.233 | 0.1523 | No |
| 26 | Kalrn | 2949 | 0.229 | 0.1576 | No |
| 27 | Cdc42bpa | 3001 | 0.225 | 0.1610 | No |
| 28 | Atp2c1 | 3021 | 0.223 | 0.1672 | No |
| 29 | Bckdhb | 3036 | 0.221 | 0.1738 | No |
| 30 | Nrp1 | 3049 | 0.221 | 0.1805 | No |
| 31 | Smad3 | 3212 | 0.209 | 0.1738 | No |
| 32 | Magi2 | 3326 | 0.198 | 0.1709 | No |
| 33 | Kit | 3442 | 0.189 | 0.1676 | No |
| 34 | Prdm2 | 3579 | 0.177 | 0.1620 | No |
| 35 | Sfmbt1 | 3632 | 0.173 | 0.1636 | No |
| 36 | Myc | 3984 | 0.143 | 0.1381 | No |
| 37 | Cdon | 4103 | 0.136 | 0.1326 | No |
| 38 | Zmiz1 | 4290 | 0.120 | 0.1207 | No |
| 39 | Runx1 | 4448 | 0.109 | 0.1109 | No |
| 40 | Fyn | 4494 | 0.106 | 0.1107 | No |
| 41 | Smad7 | 4609 | 0.097 | 0.1042 | No |
| 42 | Wdr37 | 4640 | 0.095 | 0.1050 | No |
| 43 | Pdlim5 | 4652 | 0.094 | 0.1073 | No |
| 44 | Vldlr | 4673 | 0.092 | 0.1088 | No |
| 45 | Inpp4b | 4681 | 0.092 | 0.1115 | No |
| 46 | Acvr2a | 4692 | 0.091 | 0.1138 | No |
| 47 | Itgb3 | 4992 | 0.067 | 0.0902 | No |
| 48 | Anxa4 | 5000 | 0.065 | 0.0919 | No |
| 49 | Tfpi | 5403 | 0.035 | 0.0582 | No |
| 50 | Ythdc1 | 5440 | 0.032 | 0.0561 | No |
| 51 | Plcb4 | 5751 | 0.010 | 0.0296 | No |
| 52 | Insig1 | 5852 | 0.002 | 0.0209 | No |
| 53 | Ica1 | 5871 | 0.001 | 0.0194 | No |
| 54 | Anxa2 | 6045 | -0.011 | 0.0048 | No |
| 55 | Arhgef9 | 6118 | -0.017 | -0.0009 | No |
| 56 | Nek7 | 6153 | -0.020 | -0.0031 | No |
| 57 | Tgfbr2 | 6398 | -0.037 | -0.0230 | No |
| 58 | Rxra | 6468 | -0.041 | -0.0276 | No |
| 59 | Prkce | 6589 | -0.052 | -0.0362 | No |
| 60 | Prkca | 6591 | -0.052 | -0.0344 | No |
| 61 | Phf3 | 7034 | -0.090 | -0.0697 | No |
| 62 | Pik3cd | 7110 | -0.096 | -0.0728 | No |
| 63 | Spop | 7144 | -0.098 | -0.0722 | No |
| 64 | Pias3 | 7196 | -0.101 | -0.0731 | No |
| 65 | Grk5 | 7365 | -0.113 | -0.0837 | No |
| 66 | Ldlr | 7388 | -0.116 | -0.0815 | No |
| 67 | Atp2b1 | 7396 | -0.116 | -0.0781 | No |
| 68 | Dmac2l | 7405 | -0.117 | -0.0746 | No |
| 69 | Nfkb1 | 7406 | -0.117 | -0.0705 | No |
| 70 | Gja1 | 7547 | -0.126 | -0.0782 | No |
| 71 | Pten | 7569 | -0.128 | -0.0756 | No |
| 72 | Nr1d2 | 7581 | -0.130 | -0.0719 | No |
| 73 | Ltbp1 | 7589 | -0.130 | -0.0680 | No |
| 74 | Pdgfrb | 7952 | -0.158 | -0.0939 | No |
| 75 | Mrps31 | 7970 | -0.159 | -0.0898 | No |
| 76 | Mapk14 | 8140 | -0.172 | -0.0984 | No |
| 77 | Nr3c1 | 8315 | -0.189 | -0.1068 | No |
| 78 | Scaf8 | 8327 | -0.190 | -0.1011 | No |
| 79 | Aggf1 | 8480 | -0.202 | -0.1072 | No |
| 80 | Togaram1 | 8774 | -0.231 | -0.1245 | No |
| 81 | Syne1 | 9357 | -0.286 | -0.1650 | No |
| 82 | Dlg1 | 9856 | -0.341 | -0.1963 | No |
| 83 | Tgfbr3 | 9868 | -0.343 | -0.1851 | No |
| 84 | Nipbl | 10064 | -0.365 | -0.1892 | No |
| 85 | Cdk13 | 10284 | -0.396 | -0.1943 | No |
| 86 | Mgmt | 10488 | -0.426 | -0.1970 | Yes |
| 87 | Met | 10489 | -0.427 | -0.1819 | Yes |
| 88 | Notch2 | 10531 | -0.433 | -0.1702 | Yes |
| 89 | Bmpr1a | 10548 | -0.435 | -0.1563 | Yes |
| 90 | Map2k5 | 10767 | -0.473 | -0.1586 | Yes |
| 91 | Dyrk1a | 10897 | -0.503 | -0.1521 | Yes |
| 92 | Celf2 | 11012 | -0.535 | -0.1432 | Yes |
| 93 | Lamc1 | 11013 | -0.535 | -0.1243 | Yes |
| 94 | Sipa1l1 | 11020 | -0.537 | -0.1059 | Yes |
| 95 | Atp2b4 | 11172 | -0.583 | -0.0985 | Yes |
| 96 | Cdkn1b | 11177 | -0.584 | -0.0783 | Yes |
| 97 | Akt3 | 11256 | -0.614 | -0.0635 | Yes |
| 98 | Atxn1 | 11515 | -0.816 | -0.0571 | Yes |
| 99 | Tjp1 | 11536 | -0.854 | -0.0288 | Yes |
| 100 | Dusp1 | 11579 | -0.995 | 0.0026 | Yes |