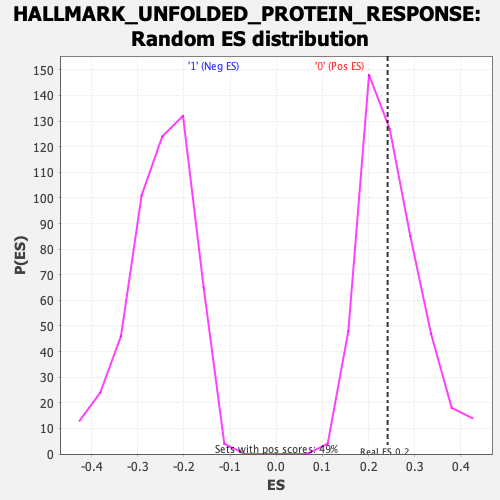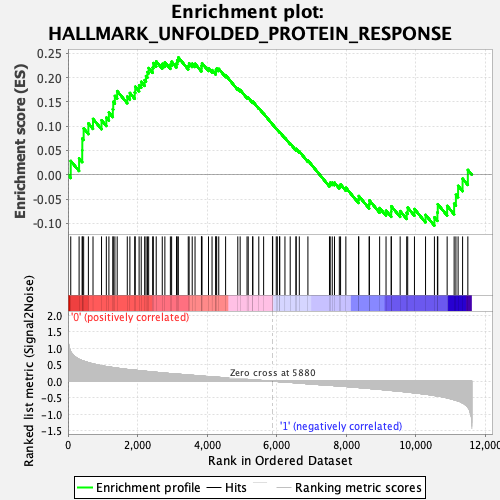
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.24132955 |
| Normalized Enrichment Score (NES) | 0.967051 |
| Nominal p-value | 0.49490836 |
| FDR q-value | 0.77953774 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nolc1 | 79 | 0.895 | 0.0283 | Yes |
| 2 | Nop56 | 319 | 0.662 | 0.0336 | Yes |
| 3 | Eif4e | 409 | 0.623 | 0.0504 | Yes |
| 4 | Psat1 | 412 | 0.621 | 0.0746 | Yes |
| 5 | Dkc1 | 450 | 0.607 | 0.0952 | Yes |
| 6 | Cnot4 | 586 | 0.562 | 0.1056 | Yes |
| 7 | Eif2ak3 | 719 | 0.525 | 0.1148 | Yes |
| 8 | Nfya | 964 | 0.469 | 0.1121 | Yes |
| 9 | Atf4 | 1102 | 0.444 | 0.1176 | Yes |
| 10 | Yif1a | 1176 | 0.433 | 0.1283 | Yes |
| 11 | Khsrp | 1289 | 0.415 | 0.1349 | Yes |
| 12 | Edc4 | 1301 | 0.413 | 0.1501 | Yes |
| 13 | Eif4a2 | 1348 | 0.407 | 0.1621 | Yes |
| 14 | Parn | 1414 | 0.396 | 0.1721 | Yes |
| 15 | Srpr | 1704 | 0.356 | 0.1610 | Yes |
| 16 | Edem1 | 1779 | 0.348 | 0.1682 | Yes |
| 17 | Serp1 | 1918 | 0.333 | 0.1693 | Yes |
| 18 | Exosc4 | 1933 | 0.332 | 0.1812 | Yes |
| 19 | Sdad1 | 2044 | 0.319 | 0.1842 | Yes |
| 20 | Calr | 2105 | 0.313 | 0.1913 | Yes |
| 21 | Pdia5 | 2204 | 0.302 | 0.1946 | Yes |
| 22 | Eif2s1 | 2243 | 0.299 | 0.2031 | Yes |
| 23 | Ero1a | 2284 | 0.294 | 0.2111 | Yes |
| 24 | Eif4a3 | 2319 | 0.289 | 0.2195 | Yes |
| 25 | Slc30a5 | 2434 | 0.277 | 0.2205 | Yes |
| 26 | Eef2 | 2453 | 0.275 | 0.2298 | Yes |
| 27 | Cebpg | 2536 | 0.269 | 0.2332 | Yes |
| 28 | Exosc9 | 2710 | 0.253 | 0.2281 | Yes |
| 29 | Zbtb17 | 2785 | 0.246 | 0.2313 | Yes |
| 30 | Nhp2 | 2943 | 0.230 | 0.2267 | Yes |
| 31 | Eif4ebp1 | 2979 | 0.227 | 0.2326 | Yes |
| 32 | Pdia6 | 3119 | 0.217 | 0.2291 | Yes |
| 33 | Nfyb | 3140 | 0.214 | 0.2357 | Yes |
| 34 | Hyou1 | 3172 | 0.211 | 0.2413 | Yes |
| 35 | Exosc2 | 3457 | 0.188 | 0.2240 | No |
| 36 | Aldh18a1 | 3482 | 0.185 | 0.2292 | No |
| 37 | Exosc1 | 3571 | 0.177 | 0.2286 | No |
| 38 | Dnajc3 | 3652 | 0.171 | 0.2283 | No |
| 39 | Cks1b | 3836 | 0.155 | 0.2186 | No |
| 40 | Arfgap1 | 3838 | 0.155 | 0.2246 | No |
| 41 | Spcs1 | 3853 | 0.154 | 0.2294 | No |
| 42 | Exosc5 | 4039 | 0.139 | 0.2188 | No |
| 43 | Gosr2 | 4141 | 0.132 | 0.2152 | No |
| 44 | Nabp1 | 4247 | 0.124 | 0.2110 | No |
| 45 | Banf1 | 4252 | 0.123 | 0.2154 | No |
| 46 | Exoc2 | 4274 | 0.121 | 0.2184 | No |
| 47 | Paip1 | 4334 | 0.117 | 0.2178 | No |
| 48 | Rrp9 | 4531 | 0.103 | 0.2049 | No |
| 49 | Dctn1 | 4881 | 0.075 | 0.1775 | No |
| 50 | Fus | 4949 | 0.070 | 0.1744 | No |
| 51 | Sec31a | 5147 | 0.055 | 0.1594 | No |
| 52 | Hspa9 | 5181 | 0.052 | 0.1586 | No |
| 53 | Hspa5 | 5308 | 0.043 | 0.1494 | No |
| 54 | Slc7a5 | 5314 | 0.042 | 0.1506 | No |
| 55 | Hsp90b1 | 5485 | 0.029 | 0.1370 | No |
| 56 | Ssr1 | 5624 | 0.018 | 0.1257 | No |
| 57 | Rps14 | 5878 | 0.000 | 0.1037 | No |
| 58 | Dnaja4 | 5884 | 0.000 | 0.1033 | No |
| 59 | Dcp2 | 5987 | -0.006 | 0.0946 | No |
| 60 | Shc1 | 6017 | -0.009 | 0.0925 | No |
| 61 | Xpot | 6081 | -0.014 | 0.0875 | No |
| 62 | Npm1 | 6088 | -0.015 | 0.0876 | No |
| 63 | Ern1 | 6237 | -0.025 | 0.0757 | No |
| 64 | Preb | 6388 | -0.036 | 0.0641 | No |
| 65 | Atp6v0d1 | 6553 | -0.050 | 0.0518 | No |
| 66 | Srprb | 6560 | -0.050 | 0.0533 | No |
| 67 | Eif4g1 | 6650 | -0.057 | 0.0478 | No |
| 68 | Dnajb9 | 6897 | -0.078 | 0.0295 | No |
| 69 | Eif4a1 | 7517 | -0.124 | -0.0194 | No |
| 70 | Cnot2 | 7538 | -0.125 | -0.0162 | No |
| 71 | Wfs1 | 7597 | -0.130 | -0.0161 | No |
| 72 | Lsm1 | 7661 | -0.134 | -0.0163 | No |
| 73 | Kif5b | 7800 | -0.144 | -0.0227 | No |
| 74 | Ddx10 | 7836 | -0.147 | -0.0199 | No |
| 75 | H2ax | 7988 | -0.160 | -0.0267 | No |
| 76 | Mthfd2 | 8352 | -0.192 | -0.0507 | No |
| 77 | Imp3 | 8360 | -0.192 | -0.0438 | No |
| 78 | Pop4 | 8658 | -0.220 | -0.0609 | No |
| 79 | Wipi1 | 8665 | -0.220 | -0.0528 | No |
| 80 | Exosc10 | 8959 | -0.247 | -0.0685 | No |
| 81 | Cxxc1 | 9143 | -0.266 | -0.0739 | No |
| 82 | Lsm4 | 9292 | -0.281 | -0.0758 | No |
| 83 | Tatdn2 | 9295 | -0.281 | -0.0649 | No |
| 84 | Sec11a | 9549 | -0.305 | -0.0749 | No |
| 85 | Asns | 9737 | -0.327 | -0.0782 | No |
| 86 | Bag3 | 9766 | -0.330 | -0.0677 | No |
| 87 | Herpud1 | 9960 | -0.353 | -0.0706 | No |
| 88 | Vegfa | 10279 | -0.395 | -0.0826 | No |
| 89 | Cnot6 | 10534 | -0.434 | -0.0877 | No |
| 90 | Mtrex | 10620 | -0.447 | -0.0775 | No |
| 91 | Ywhaz | 10631 | -0.449 | -0.0607 | No |
| 92 | Atf6 | 10900 | -0.504 | -0.0642 | No |
| 93 | Nop14 | 11101 | -0.562 | -0.0594 | No |
| 94 | Dcp1a | 11152 | -0.576 | -0.0411 | No |
| 95 | Tspyl2 | 11215 | -0.602 | -0.0229 | No |
| 96 | Xbp1 | 11344 | -0.659 | -0.0081 | No |
| 97 | Cebpb | 11495 | -0.789 | 0.0099 | No |