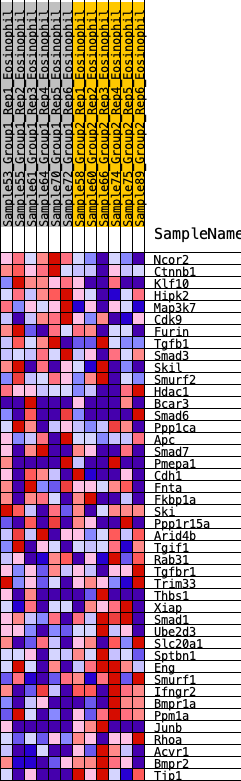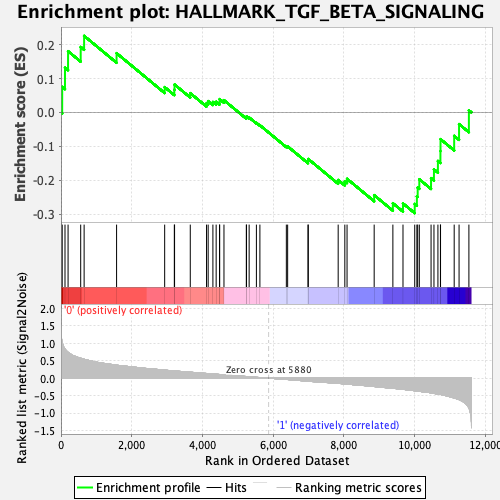
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.2967711 |
| Normalized Enrichment Score (NES) | -1.1881126 |
| Nominal p-value | 0.20958084 |
| FDR q-value | 0.44451737 |
| FWER p-Value | 0.946 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ncor2 | 34 | 1.034 | 0.0753 | No |
| 2 | Ctnnb1 | 113 | 0.837 | 0.1319 | No |
| 3 | Klf10 | 200 | 0.740 | 0.1805 | No |
| 4 | Hipk2 | 557 | 0.572 | 0.1930 | No |
| 5 | Map3k7 | 654 | 0.543 | 0.2257 | No |
| 6 | Cdk9 | 1572 | 0.373 | 0.1747 | No |
| 7 | Furin | 2932 | 0.231 | 0.0747 | No |
| 8 | Tgfb1 | 3205 | 0.209 | 0.0670 | No |
| 9 | Smad3 | 3212 | 0.209 | 0.0823 | No |
| 10 | Skil | 3656 | 0.171 | 0.0569 | No |
| 11 | Smurf2 | 4116 | 0.134 | 0.0273 | No |
| 12 | Hdac1 | 4162 | 0.131 | 0.0334 | No |
| 13 | Bcar3 | 4296 | 0.119 | 0.0309 | No |
| 14 | Smad6 | 4388 | 0.113 | 0.0315 | No |
| 15 | Ppp1ca | 4485 | 0.106 | 0.0313 | No |
| 16 | Apc | 4486 | 0.106 | 0.0393 | No |
| 17 | Smad7 | 4609 | 0.097 | 0.0361 | No |
| 18 | Pmepa1 | 5241 | 0.048 | -0.0148 | No |
| 19 | Cdh1 | 5247 | 0.047 | -0.0117 | No |
| 20 | Fnta | 5321 | 0.042 | -0.0148 | No |
| 21 | Fkbp1a | 5526 | 0.025 | -0.0306 | No |
| 22 | Ski | 5625 | 0.018 | -0.0377 | No |
| 23 | Ppp1r15a | 6373 | -0.035 | -0.0997 | No |
| 24 | Arid4b | 6410 | -0.037 | -0.1000 | No |
| 25 | Tgif1 | 6986 | -0.086 | -0.1432 | No |
| 26 | Rab31 | 6993 | -0.087 | -0.1372 | No |
| 27 | Tgfbr1 | 7839 | -0.148 | -0.1991 | No |
| 28 | Trim33 | 8027 | -0.163 | -0.2029 | No |
| 29 | Thbs1 | 8090 | -0.168 | -0.1956 | No |
| 30 | Xiap | 8857 | -0.238 | -0.2438 | No |
| 31 | Smad1 | 9386 | -0.289 | -0.2676 | No |
| 32 | Ube2d3 | 9672 | -0.318 | -0.2682 | No |
| 33 | Slc20a1 | 10004 | -0.358 | -0.2697 | Yes |
| 34 | Sptbn1 | 10065 | -0.365 | -0.2472 | Yes |
| 35 | Eng | 10088 | -0.367 | -0.2214 | Yes |
| 36 | Smurf1 | 10134 | -0.374 | -0.1970 | Yes |
| 37 | Ifngr2 | 10466 | -0.421 | -0.1937 | Yes |
| 38 | Bmpr1a | 10548 | -0.435 | -0.1678 | Yes |
| 39 | Ppm1a | 10660 | -0.454 | -0.1430 | Yes |
| 40 | Junb | 10731 | -0.466 | -0.1138 | Yes |
| 41 | Rhoa | 10734 | -0.466 | -0.0787 | Yes |
| 42 | Acvr1 | 11120 | -0.568 | -0.0690 | Yes |
| 43 | Bmpr2 | 11258 | -0.614 | -0.0344 | Yes |
| 44 | Tjp1 | 11536 | -0.854 | 0.0063 | Yes |