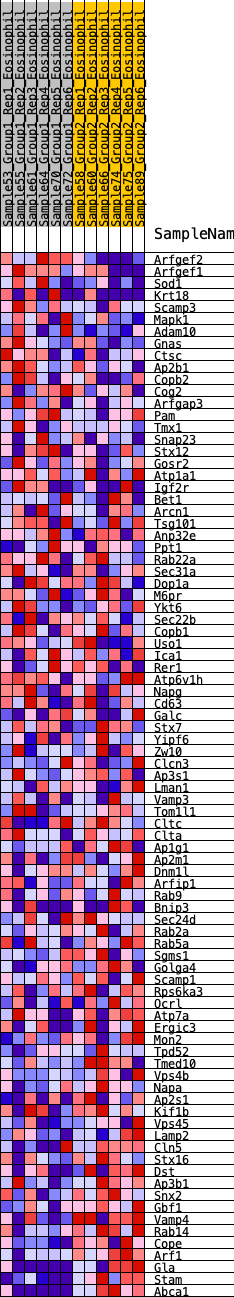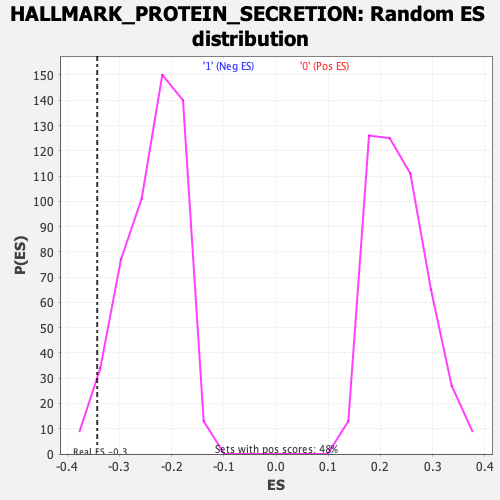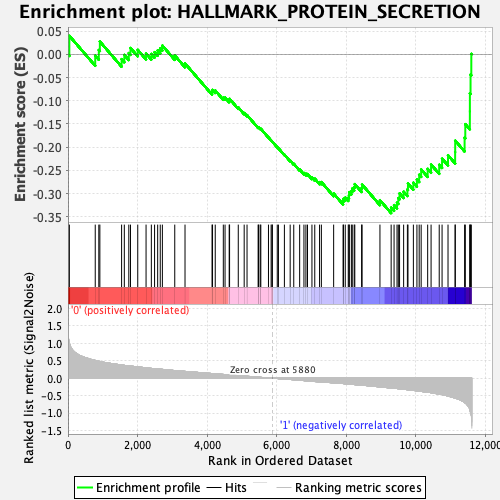
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.34222007 |
| Normalized Enrichment Score (NES) | -1.4543171 |
| Nominal p-value | 0.020992367 |
| FDR q-value | 0.18483523 |
| FWER p-Value | 0.383 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Arfgef2 | 39 | 1.007 | 0.0400 | No |
| 2 | Arfgef1 | 783 | 0.510 | -0.0025 | No |
| 3 | Sod1 | 886 | 0.487 | 0.0096 | No |
| 4 | Krt18 | 916 | 0.478 | 0.0278 | No |
| 5 | Scamp3 | 1541 | 0.377 | -0.0102 | No |
| 6 | Mapk1 | 1619 | 0.367 | -0.0010 | No |
| 7 | Adam10 | 1748 | 0.352 | 0.0030 | No |
| 8 | Gnas | 1796 | 0.347 | 0.0139 | No |
| 9 | Ctsc | 2005 | 0.325 | 0.0099 | No |
| 10 | Ap2b1 | 2244 | 0.299 | 0.0021 | No |
| 11 | Copb2 | 2396 | 0.280 | 0.0011 | No |
| 12 | Cog2 | 2489 | 0.272 | 0.0048 | No |
| 13 | Arfgap3 | 2578 | 0.265 | 0.0086 | No |
| 14 | Pam | 2652 | 0.258 | 0.0134 | No |
| 15 | Tmx1 | 2713 | 0.253 | 0.0191 | No |
| 16 | Snap23 | 3069 | 0.220 | -0.0023 | No |
| 17 | Stx12 | 3364 | 0.195 | -0.0194 | No |
| 18 | Gosr2 | 4141 | 0.132 | -0.0810 | No |
| 19 | Atp1a1 | 4155 | 0.131 | -0.0765 | No |
| 20 | Igf2r | 4233 | 0.125 | -0.0778 | No |
| 21 | Bet1 | 4465 | 0.108 | -0.0932 | No |
| 22 | Arcn1 | 4515 | 0.104 | -0.0929 | No |
| 23 | Tsg101 | 4634 | 0.096 | -0.0990 | No |
| 24 | Anp32e | 4645 | 0.095 | -0.0958 | No |
| 25 | Ppt1 | 4895 | 0.074 | -0.1143 | No |
| 26 | Rab22a | 5065 | 0.061 | -0.1263 | No |
| 27 | Sec31a | 5147 | 0.055 | -0.1310 | No |
| 28 | Dop1a | 5466 | 0.030 | -0.1573 | No |
| 29 | M6pr | 5478 | 0.029 | -0.1570 | No |
| 30 | Ykt6 | 5524 | 0.025 | -0.1598 | No |
| 31 | Sec22b | 5543 | 0.023 | -0.1604 | No |
| 32 | Copb1 | 5763 | 0.009 | -0.1790 | No |
| 33 | Uso1 | 5840 | 0.003 | -0.1855 | No |
| 34 | Ica1 | 5871 | 0.001 | -0.1880 | No |
| 35 | Rer1 | 5872 | 0.001 | -0.1880 | No |
| 36 | Atp6v1h | 6016 | -0.008 | -0.2000 | No |
| 37 | Napg | 6050 | -0.012 | -0.2024 | No |
| 38 | Cd63 | 6221 | -0.024 | -0.2161 | No |
| 39 | Galc | 6387 | -0.036 | -0.2289 | No |
| 40 | Stx7 | 6490 | -0.043 | -0.2359 | No |
| 41 | Yipf6 | 6660 | -0.058 | -0.2480 | No |
| 42 | Zw10 | 6783 | -0.068 | -0.2557 | No |
| 43 | Clcn3 | 6840 | -0.073 | -0.2574 | No |
| 44 | Ap3s1 | 6885 | -0.077 | -0.2579 | No |
| 45 | Lman1 | 7013 | -0.088 | -0.2651 | No |
| 46 | Vamp3 | 7094 | -0.095 | -0.2679 | No |
| 47 | Tom1l1 | 7232 | -0.103 | -0.2754 | No |
| 48 | Cltc | 7284 | -0.107 | -0.2752 | No |
| 49 | Clta | 7637 | -0.132 | -0.3000 | No |
| 50 | Ap1g1 | 7908 | -0.153 | -0.3169 | No |
| 51 | Ap2m1 | 7921 | -0.154 | -0.3113 | No |
| 52 | Dnm1l | 7971 | -0.159 | -0.3087 | No |
| 53 | Arfip1 | 8057 | -0.165 | -0.3090 | No |
| 54 | Rab9 | 8078 | -0.167 | -0.3035 | No |
| 55 | Bnip3 | 8087 | -0.168 | -0.2970 | No |
| 56 | Sec24d | 8146 | -0.173 | -0.2946 | No |
| 57 | Rab2a | 8172 | -0.175 | -0.2892 | No |
| 58 | Rab5a | 8227 | -0.181 | -0.2861 | No |
| 59 | Sgms1 | 8245 | -0.183 | -0.2796 | No |
| 60 | Golga4 | 8441 | -0.197 | -0.2881 | No |
| 61 | Scamp1 | 8454 | -0.199 | -0.2805 | No |
| 62 | Rps6ka3 | 8968 | -0.248 | -0.3144 | No |
| 63 | Ocrl | 9290 | -0.280 | -0.3301 | Yes |
| 64 | Atp7a | 9374 | -0.288 | -0.3249 | Yes |
| 65 | Ergic3 | 9459 | -0.296 | -0.3194 | Yes |
| 66 | Mon2 | 9496 | -0.300 | -0.3097 | Yes |
| 67 | Tpd52 | 9532 | -0.303 | -0.2996 | Yes |
| 68 | Tmed10 | 9649 | -0.316 | -0.2961 | Yes |
| 69 | Vps4b | 9757 | -0.330 | -0.2912 | Yes |
| 70 | Napa | 9774 | -0.331 | -0.2783 | Yes |
| 71 | Ap2s1 | 9929 | -0.350 | -0.2765 | Yes |
| 72 | Kif1b | 10031 | -0.362 | -0.2697 | Yes |
| 73 | Vps45 | 10096 | -0.368 | -0.2594 | Yes |
| 74 | Lamp2 | 10153 | -0.377 | -0.2480 | Yes |
| 75 | Cln5 | 10340 | -0.403 | -0.2468 | Yes |
| 76 | Stx16 | 10438 | -0.415 | -0.2373 | Yes |
| 77 | Dst | 10672 | -0.456 | -0.2379 | Yes |
| 78 | Ap3b1 | 10754 | -0.470 | -0.2247 | Yes |
| 79 | Snx2 | 10925 | -0.511 | -0.2174 | Yes |
| 80 | Gbf1 | 11130 | -0.571 | -0.2105 | Yes |
| 81 | Vamp4 | 11132 | -0.571 | -0.1859 | Yes |
| 82 | Rab14 | 11402 | -0.693 | -0.1794 | Yes |
| 83 | Cope | 11422 | -0.711 | -0.1504 | Yes |
| 84 | Arf1 | 11553 | -0.902 | -0.1228 | Yes |
| 85 | Gla | 11554 | -0.902 | -0.0839 | Yes |
| 86 | Stam | 11573 | -0.976 | -0.0434 | Yes |
| 87 | Abca1 | 11595 | -1.077 | 0.0012 | Yes |