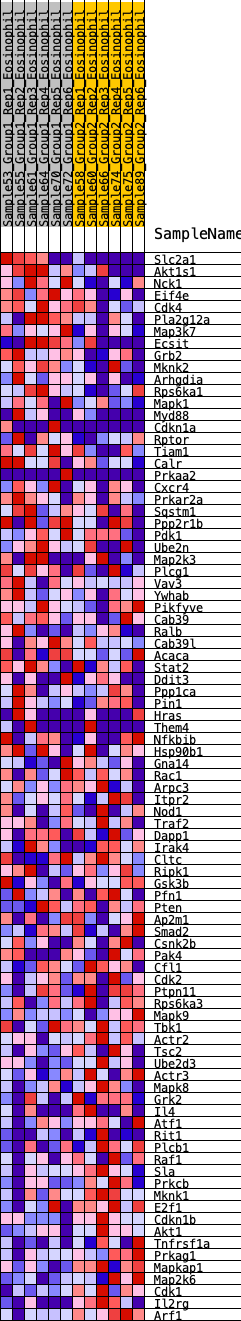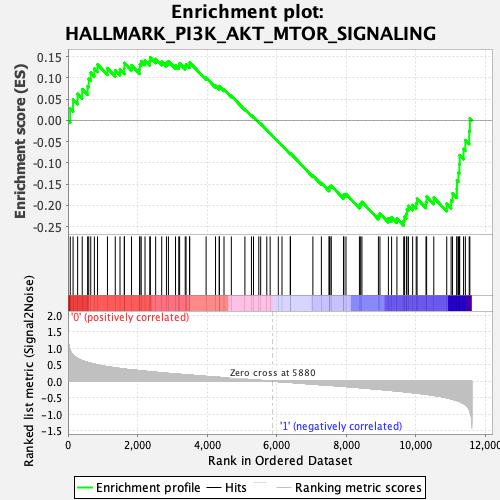
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.24785979 |
| Normalized Enrichment Score (NES) | -1.12861 |
| Nominal p-value | 0.19961241 |
| FDR q-value | 0.5003891 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc2a1 | 65 | 0.936 | 0.0278 | No |
| 2 | Akt1s1 | 146 | 0.791 | 0.0491 | No |
| 3 | Nck1 | 277 | 0.689 | 0.0624 | No |
| 4 | Eif4e | 409 | 0.623 | 0.0733 | No |
| 5 | Cdk4 | 564 | 0.571 | 0.0804 | No |
| 6 | Pla2g12a | 596 | 0.560 | 0.0977 | No |
| 7 | Map3k7 | 654 | 0.543 | 0.1121 | No |
| 8 | Ecsit | 758 | 0.516 | 0.1216 | No |
| 9 | Grb2 | 849 | 0.496 | 0.1315 | No |
| 10 | Mknk2 | 1134 | 0.439 | 0.1225 | No |
| 11 | Arhgdia | 1359 | 0.406 | 0.1176 | No |
| 12 | Rps6ka1 | 1495 | 0.383 | 0.1195 | No |
| 13 | Mapk1 | 1619 | 0.367 | 0.1220 | No |
| 14 | Myd88 | 1621 | 0.367 | 0.1350 | No |
| 15 | Cdkn1a | 1826 | 0.343 | 0.1295 | No |
| 16 | Rptor | 2060 | 0.318 | 0.1206 | No |
| 17 | Tiam1 | 2067 | 0.317 | 0.1315 | No |
| 18 | Calr | 2105 | 0.313 | 0.1394 | No |
| 19 | Prkaa2 | 2214 | 0.301 | 0.1408 | No |
| 20 | Cxcr4 | 2351 | 0.286 | 0.1392 | No |
| 21 | Prkar2a | 2367 | 0.283 | 0.1480 | No |
| 22 | Sqstm1 | 2520 | 0.270 | 0.1445 | No |
| 23 | Ppp2r1b | 2694 | 0.254 | 0.1385 | No |
| 24 | Pdk1 | 2830 | 0.241 | 0.1354 | No |
| 25 | Ube2n | 2887 | 0.236 | 0.1390 | No |
| 26 | Map2k3 | 3091 | 0.218 | 0.1292 | No |
| 27 | Plcg1 | 3185 | 0.210 | 0.1286 | No |
| 28 | Vav3 | 3209 | 0.209 | 0.1341 | No |
| 29 | Ywhab | 3370 | 0.194 | 0.1271 | No |
| 30 | Pikfyve | 3394 | 0.192 | 0.1320 | No |
| 31 | Cab39 | 3496 | 0.184 | 0.1298 | No |
| 32 | Ralb | 3498 | 0.184 | 0.1363 | No |
| 33 | Cab39l | 3970 | 0.144 | 0.1006 | No |
| 34 | Acaca | 4241 | 0.124 | 0.0816 | No |
| 35 | Stat2 | 4348 | 0.116 | 0.0765 | No |
| 36 | Ddit3 | 4351 | 0.116 | 0.0805 | No |
| 37 | Ppp1ca | 4485 | 0.106 | 0.0727 | No |
| 38 | Pin1 | 4696 | 0.091 | 0.0577 | No |
| 39 | Hras | 5086 | 0.059 | 0.0261 | No |
| 40 | Them4 | 5274 | 0.046 | 0.0114 | No |
| 41 | Nfkbib | 5333 | 0.041 | 0.0079 | No |
| 42 | Hsp90b1 | 5485 | 0.029 | -0.0042 | No |
| 43 | Gna14 | 5539 | 0.024 | -0.0080 | No |
| 44 | Rac1 | 5715 | 0.012 | -0.0227 | No |
| 45 | Arpc3 | 5812 | 0.006 | -0.0308 | No |
| 46 | Itpr2 | 6046 | -0.011 | -0.0507 | No |
| 47 | Nod1 | 6152 | -0.020 | -0.0591 | No |
| 48 | Traf2 | 6389 | -0.036 | -0.0783 | No |
| 49 | Dapp1 | 6396 | -0.036 | -0.0775 | No |
| 50 | Irak4 | 7040 | -0.090 | -0.1301 | No |
| 51 | Cltc | 7284 | -0.107 | -0.1474 | No |
| 52 | Ripk1 | 7508 | -0.123 | -0.1623 | No |
| 53 | Gsk3b | 7509 | -0.123 | -0.1579 | No |
| 54 | Pfn1 | 7532 | -0.125 | -0.1554 | No |
| 55 | Pten | 7569 | -0.128 | -0.1539 | No |
| 56 | Ap2m1 | 7921 | -0.154 | -0.1789 | No |
| 57 | Smad2 | 7932 | -0.155 | -0.1742 | No |
| 58 | Csnk2b | 7992 | -0.161 | -0.1736 | No |
| 59 | Pak4 | 8378 | -0.194 | -0.2001 | No |
| 60 | Cfl1 | 8414 | -0.196 | -0.1961 | No |
| 61 | Cdk2 | 8450 | -0.199 | -0.1920 | No |
| 62 | Ptpn11 | 8924 | -0.245 | -0.2243 | No |
| 63 | Rps6ka3 | 8968 | -0.248 | -0.2192 | No |
| 64 | Mapk9 | 9211 | -0.273 | -0.2305 | No |
| 65 | Tbk1 | 9303 | -0.282 | -0.2283 | No |
| 66 | Actr2 | 9456 | -0.296 | -0.2309 | No |
| 67 | Tsc2 | 9652 | -0.316 | -0.2366 | Yes |
| 68 | Ube2d3 | 9672 | -0.318 | -0.2269 | Yes |
| 69 | Actr3 | 9728 | -0.325 | -0.2200 | Yes |
| 70 | Mapk8 | 9744 | -0.328 | -0.2096 | Yes |
| 71 | Grk2 | 9787 | -0.334 | -0.2013 | Yes |
| 72 | Il4 | 9908 | -0.347 | -0.1994 | Yes |
| 73 | Atf1 | 10010 | -0.360 | -0.1953 | Yes |
| 74 | Rit1 | 10035 | -0.362 | -0.1844 | Yes |
| 75 | Plcb1 | 10292 | -0.397 | -0.1924 | Yes |
| 76 | Raf1 | 10310 | -0.400 | -0.1796 | Yes |
| 77 | Sla | 10516 | -0.431 | -0.1820 | Yes |
| 78 | Prkcb | 10888 | -0.501 | -0.1963 | Yes |
| 79 | Mknk1 | 11014 | -0.535 | -0.1881 | Yes |
| 80 | E2f1 | 11055 | -0.547 | -0.1720 | Yes |
| 81 | Cdkn1b | 11177 | -0.584 | -0.1616 | Yes |
| 82 | Akt1 | 11182 | -0.587 | -0.1410 | Yes |
| 83 | Tnfrsf1a | 11226 | -0.606 | -0.1231 | Yes |
| 84 | Prkag1 | 11252 | -0.612 | -0.1034 | Yes |
| 85 | Mapkap1 | 11261 | -0.615 | -0.0821 | Yes |
| 86 | Map2k6 | 11372 | -0.673 | -0.0676 | Yes |
| 87 | Cdk1 | 11424 | -0.712 | -0.0466 | Yes |
| 88 | Il2rg | 11534 | -0.847 | -0.0258 | Yes |
| 89 | Arf1 | 11553 | -0.902 | 0.0049 | Yes |