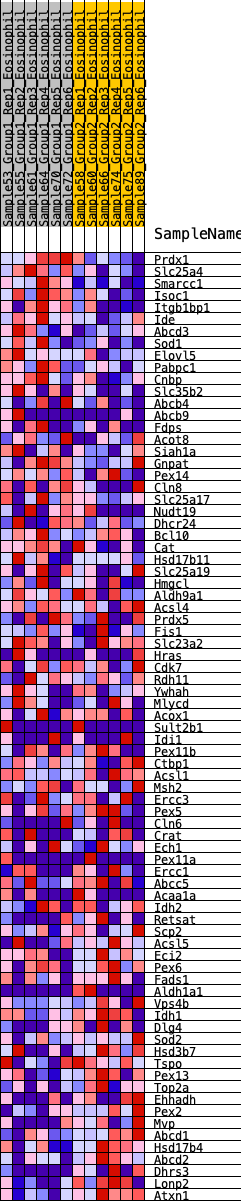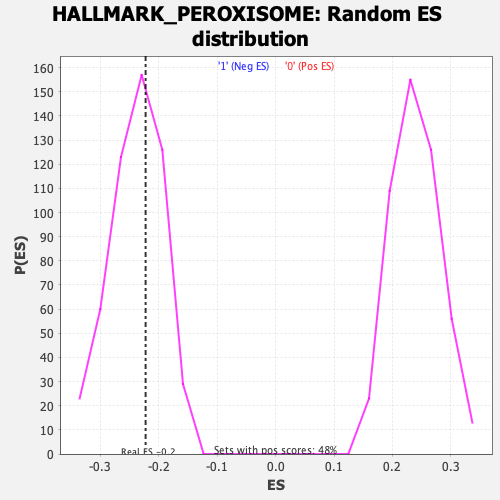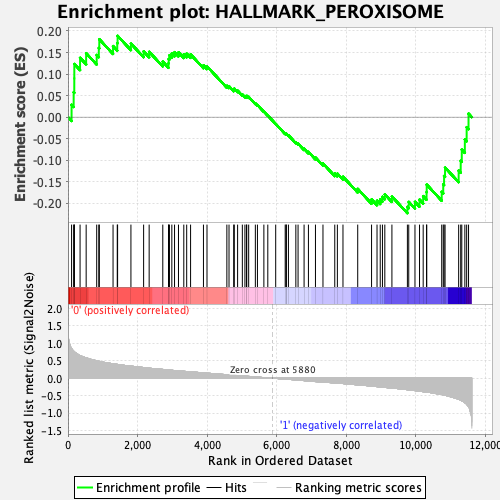
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.22268178 |
| Normalized Enrichment Score (NES) | -0.9324326 |
| Nominal p-value | 0.6003861 |
| FDR q-value | 0.6537917 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prdx1 | 103 | 0.851 | 0.0287 | No |
| 2 | Slc25a4 | 163 | 0.773 | 0.0577 | No |
| 3 | Smarcc1 | 181 | 0.759 | 0.0898 | No |
| 4 | Isoc1 | 182 | 0.758 | 0.1233 | No |
| 5 | Itgb1bp1 | 347 | 0.645 | 0.1376 | No |
| 6 | Ide | 522 | 0.582 | 0.1483 | No |
| 7 | Abcd3 | 825 | 0.501 | 0.1442 | No |
| 8 | Sod1 | 886 | 0.487 | 0.1605 | No |
| 9 | Elovl5 | 900 | 0.482 | 0.1807 | No |
| 10 | Pabpc1 | 1295 | 0.414 | 0.1648 | No |
| 11 | Cnbp | 1416 | 0.395 | 0.1719 | No |
| 12 | Slc35b2 | 1426 | 0.394 | 0.1886 | No |
| 13 | Abcb4 | 1809 | 0.345 | 0.1707 | No |
| 14 | Abcb9 | 2174 | 0.304 | 0.1526 | No |
| 15 | Fdps | 2333 | 0.288 | 0.1516 | No |
| 16 | Acot8 | 2725 | 0.251 | 0.1288 | No |
| 17 | Siah1a | 2890 | 0.236 | 0.1249 | No |
| 18 | Gnpat | 2895 | 0.235 | 0.1350 | No |
| 19 | Pex14 | 2916 | 0.233 | 0.1436 | No |
| 20 | Cln8 | 2985 | 0.226 | 0.1476 | No |
| 21 | Slc25a17 | 3063 | 0.220 | 0.1507 | No |
| 22 | Nudt19 | 3176 | 0.211 | 0.1503 | No |
| 23 | Dhcr24 | 3329 | 0.197 | 0.1459 | No |
| 24 | Bcl10 | 3412 | 0.191 | 0.1472 | No |
| 25 | Cat | 3524 | 0.182 | 0.1456 | No |
| 26 | Hsd17b11 | 3894 | 0.150 | 0.1203 | No |
| 27 | Slc25a19 | 3996 | 0.141 | 0.1177 | No |
| 28 | Hmgcl | 4567 | 0.101 | 0.0728 | No |
| 29 | Aldh9a1 | 4629 | 0.096 | 0.0717 | No |
| 30 | Acsl4 | 4764 | 0.086 | 0.0639 | No |
| 31 | Prdx5 | 4780 | 0.084 | 0.0663 | No |
| 32 | Fis1 | 4876 | 0.075 | 0.0614 | No |
| 33 | Slc23a2 | 5012 | 0.065 | 0.0526 | No |
| 34 | Hras | 5086 | 0.059 | 0.0488 | No |
| 35 | Cdk7 | 5131 | 0.056 | 0.0475 | No |
| 36 | Rdh11 | 5139 | 0.056 | 0.0494 | No |
| 37 | Ywhah | 5194 | 0.051 | 0.0469 | No |
| 38 | Mlycd | 5386 | 0.036 | 0.0320 | No |
| 39 | Acox1 | 5446 | 0.031 | 0.0282 | No |
| 40 | Sult2b1 | 5631 | 0.017 | 0.0130 | No |
| 41 | Idi1 | 5744 | 0.010 | 0.0037 | No |
| 42 | Pex11b | 5971 | -0.005 | -0.0156 | No |
| 43 | Ctbp1 | 6244 | -0.026 | -0.0381 | No |
| 44 | Acsl1 | 6270 | -0.027 | -0.0391 | No |
| 45 | Msh2 | 6278 | -0.028 | -0.0384 | No |
| 46 | Ercc3 | 6337 | -0.032 | -0.0421 | No |
| 47 | Pex5 | 6550 | -0.049 | -0.0583 | No |
| 48 | Cln6 | 6614 | -0.054 | -0.0613 | No |
| 49 | Crat | 6787 | -0.069 | -0.0732 | No |
| 50 | Ech1 | 6912 | -0.079 | -0.0805 | No |
| 51 | Pex11a | 7114 | -0.096 | -0.0937 | No |
| 52 | Ercc1 | 7330 | -0.110 | -0.1075 | No |
| 53 | Abcc5 | 7669 | -0.135 | -0.1308 | No |
| 54 | Acaa1a | 7744 | -0.140 | -0.1310 | No |
| 55 | Idh2 | 7905 | -0.153 | -0.1381 | No |
| 56 | Retsat | 8329 | -0.190 | -0.1664 | No |
| 57 | Scp2 | 8727 | -0.226 | -0.1909 | No |
| 58 | Acsl5 | 8884 | -0.241 | -0.1937 | No |
| 59 | Eci2 | 8976 | -0.249 | -0.1906 | No |
| 60 | Pex6 | 9040 | -0.254 | -0.1848 | No |
| 61 | Fads1 | 9108 | -0.262 | -0.1791 | No |
| 62 | Aldh1a1 | 9312 | -0.283 | -0.1842 | No |
| 63 | Vps4b | 9757 | -0.330 | -0.2081 | Yes |
| 64 | Idh1 | 9796 | -0.334 | -0.1966 | Yes |
| 65 | Dlg4 | 9975 | -0.355 | -0.1964 | Yes |
| 66 | Sod2 | 10106 | -0.370 | -0.1913 | Yes |
| 67 | Hsd3b7 | 10211 | -0.386 | -0.1832 | Yes |
| 68 | Tspo | 10308 | -0.400 | -0.1739 | Yes |
| 69 | Pex13 | 10313 | -0.400 | -0.1566 | Yes |
| 70 | Top2a | 10744 | -0.468 | -0.1732 | Yes |
| 71 | Ehhadh | 10788 | -0.478 | -0.1558 | Yes |
| 72 | Pex2 | 10812 | -0.484 | -0.1364 | Yes |
| 73 | Mvp | 10840 | -0.491 | -0.1170 | Yes |
| 74 | Abcd1 | 11231 | -0.607 | -0.1240 | Yes |
| 75 | Hsd17b4 | 11290 | -0.628 | -0.1012 | Yes |
| 76 | Abcd2 | 11321 | -0.646 | -0.0753 | Yes |
| 77 | Dhrs3 | 11409 | -0.701 | -0.0518 | Yes |
| 78 | Lonp2 | 11461 | -0.744 | -0.0233 | Yes |
| 79 | Atxn1 | 11515 | -0.816 | 0.0082 | Yes |