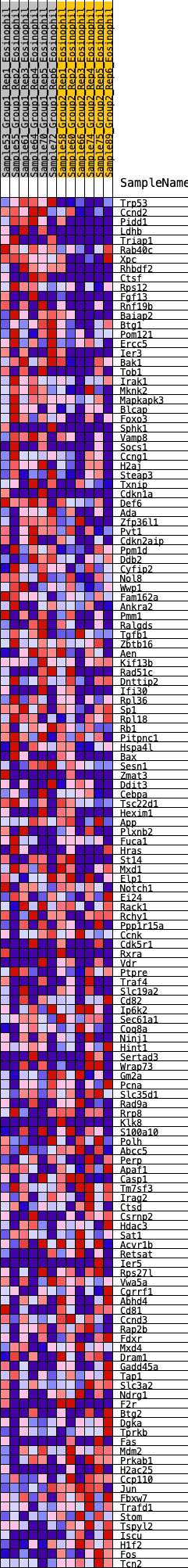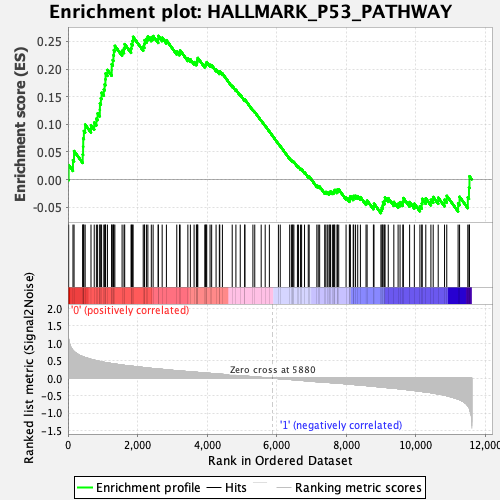
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_P53_PATHWAY |
| Enrichment Score (ES) | 0.26015404 |
| Normalized Enrichment Score (NES) | 1.1837783 |
| Nominal p-value | 0.15830116 |
| FDR q-value | 0.62901396 |
| FWER p-Value | 0.929 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Trp53 | 19 | 1.120 | 0.0261 | Yes |
| 2 | Ccnd2 | 143 | 0.795 | 0.0352 | Yes |
| 3 | Pidd1 | 174 | 0.765 | 0.0516 | Yes |
| 4 | Ldhb | 423 | 0.615 | 0.0452 | Yes |
| 5 | Triap1 | 434 | 0.613 | 0.0595 | Yes |
| 6 | Rab40c | 438 | 0.612 | 0.0745 | Yes |
| 7 | Xpc | 457 | 0.606 | 0.0879 | Yes |
| 8 | Rhbdf2 | 493 | 0.590 | 0.0995 | Yes |
| 9 | Ctsf | 661 | 0.540 | 0.0984 | Yes |
| 10 | Rps12 | 754 | 0.517 | 0.1032 | Yes |
| 11 | Fgf13 | 816 | 0.503 | 0.1104 | Yes |
| 12 | Rnf19b | 851 | 0.495 | 0.1197 | Yes |
| 13 | Baiap2 | 915 | 0.478 | 0.1261 | Yes |
| 14 | Btg1 | 918 | 0.478 | 0.1378 | Yes |
| 15 | Pom121 | 945 | 0.472 | 0.1472 | Yes |
| 16 | Ercc5 | 966 | 0.469 | 0.1571 | Yes |
| 17 | Ier3 | 1030 | 0.455 | 0.1629 | Yes |
| 18 | Bak1 | 1053 | 0.451 | 0.1722 | Yes |
| 19 | Tob1 | 1075 | 0.447 | 0.1815 | Yes |
| 20 | Irak1 | 1081 | 0.446 | 0.1921 | Yes |
| 21 | Mknk2 | 1134 | 0.439 | 0.1985 | Yes |
| 22 | Mapkapk3 | 1256 | 0.419 | 0.1984 | Yes |
| 23 | Blcap | 1257 | 0.419 | 0.2088 | Yes |
| 24 | Foxo3 | 1288 | 0.415 | 0.2164 | Yes |
| 25 | Sphk1 | 1307 | 0.412 | 0.2251 | Yes |
| 26 | Vamp8 | 1319 | 0.411 | 0.2343 | Yes |
| 27 | Socs1 | 1347 | 0.408 | 0.2421 | Yes |
| 28 | Ccng1 | 1555 | 0.374 | 0.2334 | Yes |
| 29 | H2aj | 1610 | 0.368 | 0.2378 | Yes |
| 30 | Steap3 | 1630 | 0.366 | 0.2452 | Yes |
| 31 | Txnip | 1813 | 0.344 | 0.2379 | Yes |
| 32 | Cdkn1a | 1826 | 0.343 | 0.2454 | Yes |
| 33 | Def6 | 1857 | 0.340 | 0.2512 | Yes |
| 34 | Ada | 1871 | 0.339 | 0.2585 | Yes |
| 35 | Zfp36l1 | 2162 | 0.306 | 0.2408 | Yes |
| 36 | Pvt1 | 2194 | 0.303 | 0.2456 | Yes |
| 37 | Cdkn2aip | 2203 | 0.302 | 0.2524 | Yes |
| 38 | Ppm1d | 2259 | 0.297 | 0.2550 | Yes |
| 39 | Ddb2 | 2296 | 0.292 | 0.2591 | Yes |
| 40 | Cyfip2 | 2395 | 0.281 | 0.2575 | Yes |
| 41 | Nol8 | 2447 | 0.276 | 0.2599 | Yes |
| 42 | Wwp1 | 2593 | 0.263 | 0.2538 | Yes |
| 43 | Fam162a | 2596 | 0.263 | 0.2602 | Yes |
| 44 | Ankra2 | 2707 | 0.253 | 0.2568 | No |
| 45 | Pmm1 | 2829 | 0.242 | 0.2523 | No |
| 46 | Ralgds | 3127 | 0.216 | 0.2317 | No |
| 47 | Tgfb1 | 3205 | 0.209 | 0.2302 | No |
| 48 | Zbtb16 | 3223 | 0.208 | 0.2339 | No |
| 49 | Aen | 3446 | 0.189 | 0.2192 | No |
| 50 | Kif13b | 3518 | 0.182 | 0.2175 | No |
| 51 | Rad51c | 3625 | 0.174 | 0.2126 | No |
| 52 | Dnttip2 | 3691 | 0.168 | 0.2111 | No |
| 53 | Ifi30 | 3707 | 0.167 | 0.2140 | No |
| 54 | Rpl36 | 3718 | 0.166 | 0.2172 | No |
| 55 | Sp1 | 3732 | 0.165 | 0.2202 | No |
| 56 | Rpl18 | 3933 | 0.147 | 0.2064 | No |
| 57 | Rb1 | 3955 | 0.145 | 0.2082 | No |
| 58 | Pitpnc1 | 3971 | 0.144 | 0.2104 | No |
| 59 | Hspa4l | 3988 | 0.142 | 0.2126 | No |
| 60 | Bax | 4084 | 0.137 | 0.2077 | No |
| 61 | Sesn1 | 4130 | 0.133 | 0.2071 | No |
| 62 | Zmat3 | 4257 | 0.122 | 0.1991 | No |
| 63 | Ddit3 | 4351 | 0.116 | 0.1939 | No |
| 64 | Cebpa | 4363 | 0.114 | 0.1958 | No |
| 65 | Tsc22d1 | 4439 | 0.109 | 0.1919 | No |
| 66 | Hexim1 | 4721 | 0.089 | 0.1696 | No |
| 67 | App | 4827 | 0.079 | 0.1624 | No |
| 68 | Plxnb2 | 4952 | 0.069 | 0.1534 | No |
| 69 | Fuca1 | 5079 | 0.060 | 0.1438 | No |
| 70 | Hras | 5086 | 0.059 | 0.1448 | No |
| 71 | St14 | 5316 | 0.042 | 0.1259 | No |
| 72 | Mxd1 | 5369 | 0.038 | 0.1223 | No |
| 73 | Elp1 | 5554 | 0.022 | 0.1068 | No |
| 74 | Notch1 | 5669 | 0.016 | 0.0972 | No |
| 75 | Ei24 | 5789 | 0.007 | 0.0870 | No |
| 76 | Rack1 | 6049 | -0.011 | 0.0647 | No |
| 77 | Rchy1 | 6102 | -0.016 | 0.0606 | No |
| 78 | Ppp1r15a | 6373 | -0.035 | 0.0379 | No |
| 79 | Ccnk | 6422 | -0.038 | 0.0347 | No |
| 80 | Cdk5r1 | 6450 | -0.040 | 0.0333 | No |
| 81 | Rxra | 6468 | -0.041 | 0.0328 | No |
| 82 | Vdr | 6494 | -0.044 | 0.0317 | No |
| 83 | Ptpre | 6596 | -0.052 | 0.0242 | No |
| 84 | Traf4 | 6628 | -0.056 | 0.0229 | No |
| 85 | Slc19a2 | 6686 | -0.060 | 0.0194 | No |
| 86 | Cd82 | 6716 | -0.063 | 0.0185 | No |
| 87 | Ip6k2 | 6800 | -0.070 | 0.0130 | No |
| 88 | Sec61a1 | 6907 | -0.079 | 0.0057 | No |
| 89 | Coq8a | 6933 | -0.082 | 0.0055 | No |
| 90 | Ninj1 | 7154 | -0.098 | -0.0112 | No |
| 91 | Hint1 | 7194 | -0.101 | -0.0121 | No |
| 92 | Sertad3 | 7235 | -0.104 | -0.0130 | No |
| 93 | Wrap73 | 7380 | -0.115 | -0.0227 | No |
| 94 | Gm2a | 7412 | -0.118 | -0.0225 | No |
| 95 | Pcna | 7455 | -0.119 | -0.0232 | No |
| 96 | Slc35d1 | 7503 | -0.122 | -0.0243 | No |
| 97 | Rad9a | 7526 | -0.124 | -0.0231 | No |
| 98 | Rrp8 | 7545 | -0.126 | -0.0216 | No |
| 99 | Klk8 | 7600 | -0.130 | -0.0231 | No |
| 100 | S100a10 | 7635 | -0.132 | -0.0227 | No |
| 101 | Polh | 7652 | -0.133 | -0.0208 | No |
| 102 | Abcc5 | 7669 | -0.135 | -0.0189 | No |
| 103 | Perp | 7736 | -0.140 | -0.0212 | No |
| 104 | Apaf1 | 7743 | -0.140 | -0.0182 | No |
| 105 | Casp1 | 7783 | -0.143 | -0.0181 | No |
| 106 | Tm7sf3 | 7994 | -0.161 | -0.0324 | No |
| 107 | Irag2 | 8089 | -0.168 | -0.0364 | No |
| 108 | Ctsd | 8100 | -0.168 | -0.0331 | No |
| 109 | Csrnp2 | 8118 | -0.170 | -0.0304 | No |
| 110 | Hdac3 | 8199 | -0.178 | -0.0329 | No |
| 111 | Sat1 | 8214 | -0.180 | -0.0297 | No |
| 112 | Acvr1b | 8267 | -0.185 | -0.0296 | No |
| 113 | Retsat | 8329 | -0.190 | -0.0302 | No |
| 114 | Ier5 | 8406 | -0.196 | -0.0320 | No |
| 115 | Rps27l | 8568 | -0.210 | -0.0408 | No |
| 116 | Vwa5a | 8599 | -0.213 | -0.0381 | No |
| 117 | Cgrrf1 | 8780 | -0.231 | -0.0481 | No |
| 118 | Abhd4 | 8797 | -0.232 | -0.0437 | No |
| 119 | Cd81 | 8996 | -0.250 | -0.0548 | No |
| 120 | Ccnd3 | 9024 | -0.253 | -0.0508 | No |
| 121 | Rap2b | 9050 | -0.255 | -0.0467 | No |
| 122 | Fdxr | 9056 | -0.255 | -0.0408 | No |
| 123 | Mxd4 | 9104 | -0.262 | -0.0384 | No |
| 124 | Dram1 | 9112 | -0.262 | -0.0325 | No |
| 125 | Gadd45a | 9207 | -0.273 | -0.0339 | No |
| 126 | Tap1 | 9367 | -0.287 | -0.0406 | No |
| 127 | Slc3a2 | 9490 | -0.299 | -0.0439 | No |
| 128 | Ndrg1 | 9546 | -0.305 | -0.0411 | No |
| 129 | F2r | 9626 | -0.314 | -0.0402 | No |
| 130 | Btg2 | 9641 | -0.315 | -0.0336 | No |
| 131 | Dgka | 9821 | -0.338 | -0.0408 | No |
| 132 | Tprkb | 9957 | -0.353 | -0.0438 | No |
| 133 | Fas | 10117 | -0.371 | -0.0484 | No |
| 134 | Mdm2 | 10170 | -0.380 | -0.0435 | No |
| 135 | Prkab1 | 10182 | -0.382 | -0.0350 | No |
| 136 | H2ac25 | 10286 | -0.396 | -0.0342 | No |
| 137 | Ccp110 | 10433 | -0.415 | -0.0366 | No |
| 138 | Jun | 10499 | -0.428 | -0.0316 | No |
| 139 | Fbxw7 | 10643 | -0.452 | -0.0329 | No |
| 140 | Trafd1 | 10825 | -0.487 | -0.0366 | No |
| 141 | Stom | 10889 | -0.501 | -0.0296 | No |
| 142 | Tspyl2 | 11215 | -0.602 | -0.0430 | No |
| 143 | Iscu | 11257 | -0.614 | -0.0314 | No |
| 144 | H1f2 | 11494 | -0.786 | -0.0324 | No |
| 145 | Fos | 11528 | -0.831 | -0.0146 | No |
| 146 | Tcn2 | 11542 | -0.871 | 0.0058 | No |