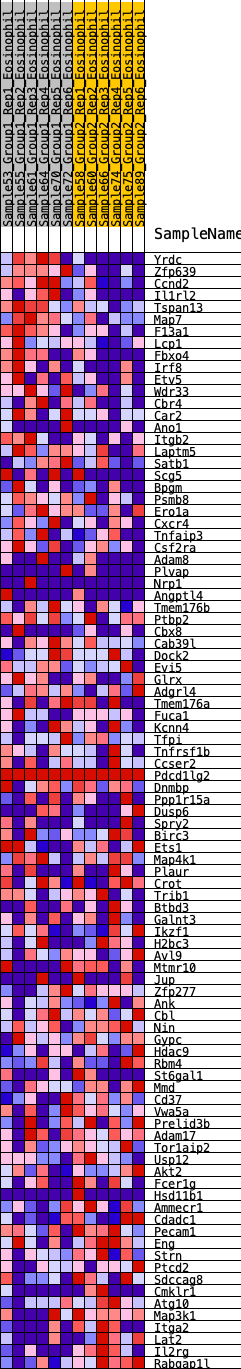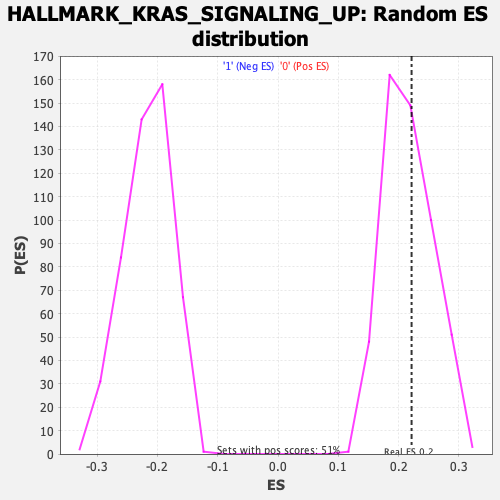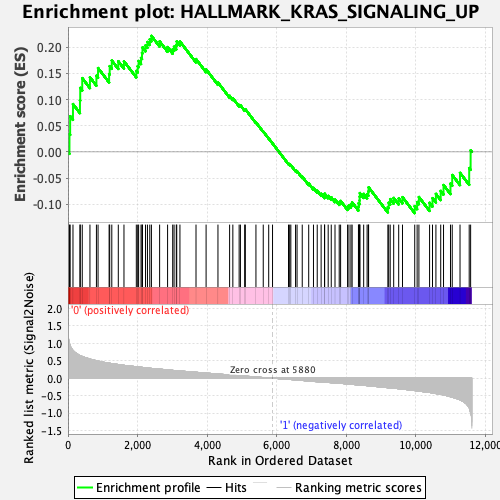
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.22145069 |
| Normalized Enrichment Score (NES) | 1.0230832 |
| Nominal p-value | 0.40272373 |
| FDR q-value | 0.99528825 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Yrdc | 42 | 0.996 | 0.0341 | Yes |
| 2 | Zfp639 | 62 | 0.942 | 0.0681 | Yes |
| 3 | Ccnd2 | 143 | 0.795 | 0.0913 | Yes |
| 4 | Il1rl2 | 344 | 0.647 | 0.0984 | Yes |
| 5 | Tspan13 | 352 | 0.644 | 0.1222 | Yes |
| 6 | Map7 | 410 | 0.621 | 0.1408 | Yes |
| 7 | F13a1 | 631 | 0.548 | 0.1424 | Yes |
| 8 | Lcp1 | 817 | 0.503 | 0.1454 | Yes |
| 9 | Fbxo4 | 864 | 0.492 | 0.1600 | Yes |
| 10 | Irf8 | 1182 | 0.431 | 0.1488 | Yes |
| 11 | Etv5 | 1199 | 0.428 | 0.1636 | Yes |
| 12 | Wdr33 | 1259 | 0.419 | 0.1744 | Yes |
| 13 | Cbr4 | 1446 | 0.390 | 0.1730 | Yes |
| 14 | Car2 | 1609 | 0.368 | 0.1729 | Yes |
| 15 | Ano1 | 1964 | 0.329 | 0.1546 | Yes |
| 16 | Itgb2 | 2004 | 0.325 | 0.1635 | Yes |
| 17 | Laptm5 | 2030 | 0.321 | 0.1735 | Yes |
| 18 | Satb1 | 2100 | 0.314 | 0.1794 | Yes |
| 19 | Scg5 | 2128 | 0.310 | 0.1888 | Yes |
| 20 | Bpgm | 2142 | 0.309 | 0.1994 | Yes |
| 21 | Psmb8 | 2229 | 0.300 | 0.2033 | Yes |
| 22 | Ero1a | 2284 | 0.294 | 0.2097 | Yes |
| 23 | Cxcr4 | 2351 | 0.286 | 0.2148 | Yes |
| 24 | Tnfaip3 | 2398 | 0.280 | 0.2215 | Yes |
| 25 | Csf2ra | 2634 | 0.260 | 0.2109 | No |
| 26 | Adam8 | 2863 | 0.237 | 0.2001 | No |
| 27 | Plvap | 3010 | 0.224 | 0.1959 | No |
| 28 | Nrp1 | 3049 | 0.221 | 0.2009 | No |
| 29 | Angptl4 | 3113 | 0.217 | 0.2037 | No |
| 30 | Tmem176b | 3126 | 0.216 | 0.2108 | No |
| 31 | Ptbp2 | 3218 | 0.208 | 0.2108 | No |
| 32 | Cbx8 | 3680 | 0.169 | 0.1771 | No |
| 33 | Cab39l | 3970 | 0.144 | 0.1575 | No |
| 34 | Dock2 | 4311 | 0.118 | 0.1325 | No |
| 35 | Evi5 | 4646 | 0.094 | 0.1070 | No |
| 36 | Glrx | 4739 | 0.088 | 0.1024 | No |
| 37 | Adgrl4 | 4920 | 0.072 | 0.0895 | No |
| 38 | Tmem176a | 4957 | 0.069 | 0.0890 | No |
| 39 | Fuca1 | 5079 | 0.060 | 0.0807 | No |
| 40 | Kcnn4 | 5098 | 0.059 | 0.0814 | No |
| 41 | Tfpi | 5403 | 0.035 | 0.0563 | No |
| 42 | Tnfrsf1b | 5613 | 0.018 | 0.0389 | No |
| 43 | Ccser2 | 5770 | 0.008 | 0.0256 | No |
| 44 | Pdcd1lg2 | 5880 | 0.000 | 0.0162 | No |
| 45 | Dnmbp | 6339 | -0.032 | -0.0224 | No |
| 46 | Ppp1r15a | 6373 | -0.035 | -0.0239 | No |
| 47 | Dusp6 | 6406 | -0.037 | -0.0253 | No |
| 48 | Spry2 | 6542 | -0.049 | -0.0352 | No |
| 49 | Birc3 | 6585 | -0.052 | -0.0369 | No |
| 50 | Ets1 | 6734 | -0.065 | -0.0473 | No |
| 51 | Map4k1 | 6920 | -0.080 | -0.0603 | No |
| 52 | Plaur | 7057 | -0.091 | -0.0687 | No |
| 53 | Crot | 7165 | -0.099 | -0.0742 | No |
| 54 | Trib1 | 7277 | -0.106 | -0.0798 | No |
| 55 | Btbd3 | 7375 | -0.115 | -0.0839 | No |
| 56 | Galnt3 | 7378 | -0.115 | -0.0797 | No |
| 57 | Ikzf1 | 7480 | -0.121 | -0.0839 | No |
| 58 | H2bc3 | 7568 | -0.128 | -0.0867 | No |
| 59 | Avl9 | 7676 | -0.135 | -0.0908 | No |
| 60 | Mtmr10 | 7797 | -0.144 | -0.0958 | No |
| 61 | Jup | 7837 | -0.148 | -0.0936 | No |
| 62 | Zfp277 | 8037 | -0.164 | -0.1047 | No |
| 63 | Ank | 8077 | -0.167 | -0.1017 | No |
| 64 | Cbl | 8134 | -0.172 | -0.1001 | No |
| 65 | Nin | 8168 | -0.175 | -0.0963 | No |
| 66 | Gypc | 8350 | -0.191 | -0.1048 | No |
| 67 | Hdac9 | 8356 | -0.192 | -0.0979 | No |
| 68 | Rbm4 | 8382 | -0.195 | -0.0927 | No |
| 69 | St6gal1 | 8385 | -0.195 | -0.0855 | No |
| 70 | Mmd | 8391 | -0.196 | -0.0785 | No |
| 71 | Cd37 | 8500 | -0.204 | -0.0802 | No |
| 72 | Vwa5a | 8599 | -0.213 | -0.0806 | No |
| 73 | Prelid3b | 8636 | -0.218 | -0.0755 | No |
| 74 | Adam17 | 8642 | -0.218 | -0.0677 | No |
| 75 | Tor1aip2 | 9196 | -0.272 | -0.1054 | No |
| 76 | Usp12 | 9220 | -0.274 | -0.0970 | No |
| 77 | Akt2 | 9263 | -0.278 | -0.0901 | No |
| 78 | Fcer1g | 9363 | -0.287 | -0.0879 | No |
| 79 | Hsd11b1 | 9507 | -0.301 | -0.0889 | No |
| 80 | Ammecr1 | 9616 | -0.313 | -0.0864 | No |
| 81 | Cdadc1 | 9968 | -0.355 | -0.1035 | No |
| 82 | Pecam1 | 10037 | -0.362 | -0.0956 | No |
| 83 | Eng | 10088 | -0.367 | -0.0861 | No |
| 84 | Strn | 10395 | -0.409 | -0.0972 | No |
| 85 | Ptcd2 | 10479 | -0.423 | -0.0884 | No |
| 86 | Sdccag8 | 10577 | -0.440 | -0.0801 | No |
| 87 | Cmklr1 | 10715 | -0.463 | -0.0745 | No |
| 88 | Atg10 | 10796 | -0.480 | -0.0632 | No |
| 89 | Map3k1 | 10996 | -0.530 | -0.0604 | No |
| 90 | Itga2 | 11046 | -0.544 | -0.0441 | No |
| 91 | Lat2 | 11270 | -0.619 | -0.0400 | No |
| 92 | Il2rg | 11534 | -0.847 | -0.0308 | No |
| 93 | Rabgap1l | 11575 | -0.982 | 0.0030 | No |