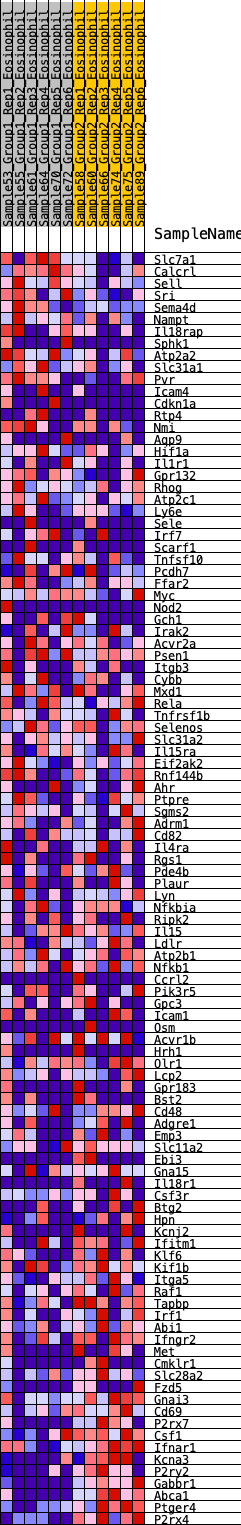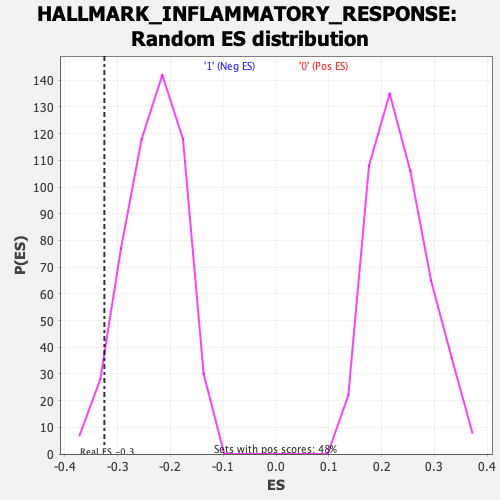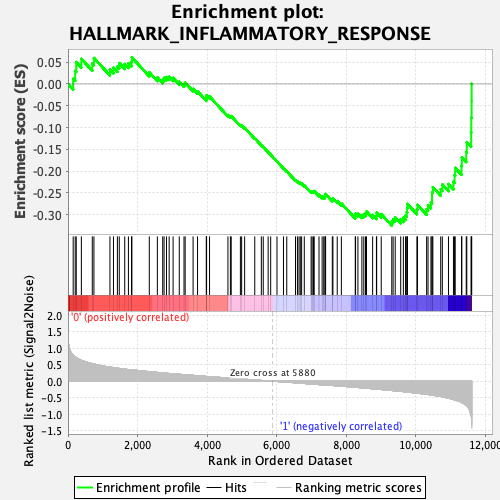
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.3251061 |
| Normalized Enrichment Score (NES) | -1.4115876 |
| Nominal p-value | 0.042307694 |
| FDR q-value | 0.20293148 |
| FWER p-Value | 0.491 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc7a1 | 149 | 0.788 | 0.0119 | No |
| 2 | Calcrl | 207 | 0.736 | 0.0302 | No |
| 3 | Sell | 237 | 0.715 | 0.0503 | No |
| 4 | Sri | 384 | 0.635 | 0.0577 | No |
| 5 | Sema4d | 698 | 0.530 | 0.0473 | No |
| 6 | Nampt | 745 | 0.519 | 0.0597 | No |
| 7 | Il18rap | 1206 | 0.427 | 0.0332 | No |
| 8 | Sphk1 | 1307 | 0.412 | 0.0375 | No |
| 9 | Atp2a2 | 1420 | 0.395 | 0.0403 | No |
| 10 | Slc31a1 | 1474 | 0.386 | 0.0479 | No |
| 11 | Pvr | 1631 | 0.366 | 0.0459 | No |
| 12 | Icam4 | 1737 | 0.353 | 0.0479 | No |
| 13 | Cdkn1a | 1826 | 0.343 | 0.0511 | No |
| 14 | Rtp4 | 1837 | 0.342 | 0.0610 | No |
| 15 | Nmi | 2337 | 0.287 | 0.0267 | No |
| 16 | Aqp9 | 2569 | 0.266 | 0.0150 | No |
| 17 | Hif1a | 2723 | 0.251 | 0.0097 | No |
| 18 | Il1r1 | 2762 | 0.247 | 0.0142 | No |
| 19 | Gpr132 | 2832 | 0.241 | 0.0158 | No |
| 20 | Rhog | 2908 | 0.234 | 0.0167 | No |
| 21 | Atp2c1 | 3021 | 0.223 | 0.0140 | No |
| 22 | Ly6e | 3199 | 0.210 | 0.0052 | No |
| 23 | Sele | 3335 | 0.197 | -0.0003 | No |
| 24 | Irf7 | 3368 | 0.194 | 0.0031 | No |
| 25 | Scarf1 | 3596 | 0.176 | -0.0111 | No |
| 26 | Tnfsf10 | 3724 | 0.166 | -0.0169 | No |
| 27 | Pcdh7 | 3976 | 0.144 | -0.0342 | No |
| 28 | Ffar2 | 3981 | 0.143 | -0.0300 | No |
| 29 | Myc | 3984 | 0.143 | -0.0257 | No |
| 30 | Nod2 | 4069 | 0.138 | -0.0286 | No |
| 31 | Gch1 | 4598 | 0.098 | -0.0714 | No |
| 32 | Irak2 | 4669 | 0.093 | -0.0746 | No |
| 33 | Acvr2a | 4692 | 0.091 | -0.0736 | No |
| 34 | Psen1 | 4956 | 0.069 | -0.0943 | No |
| 35 | Itgb3 | 4992 | 0.067 | -0.0952 | No |
| 36 | Cybb | 5078 | 0.060 | -0.1007 | No |
| 37 | Mxd1 | 5369 | 0.038 | -0.1247 | No |
| 38 | Rela | 5558 | 0.022 | -0.1404 | No |
| 39 | Tnfrsf1b | 5613 | 0.018 | -0.1445 | No |
| 40 | Selenos | 5754 | 0.009 | -0.1563 | No |
| 41 | Slc31a2 | 5827 | 0.004 | -0.1625 | No |
| 42 | Il15ra | 6008 | -0.008 | -0.1779 | No |
| 43 | Eif2ak2 | 6195 | -0.022 | -0.1933 | No |
| 44 | Rnf144b | 6289 | -0.029 | -0.2005 | No |
| 45 | Ahr | 6540 | -0.049 | -0.2207 | No |
| 46 | Ptpre | 6596 | -0.052 | -0.2238 | No |
| 47 | Sgms2 | 6641 | -0.057 | -0.2259 | No |
| 48 | Adrm1 | 6678 | -0.059 | -0.2271 | No |
| 49 | Cd82 | 6716 | -0.063 | -0.2283 | No |
| 50 | Il4ra | 6793 | -0.069 | -0.2328 | No |
| 51 | Rgs1 | 6990 | -0.086 | -0.2471 | No |
| 52 | Pde4b | 7019 | -0.088 | -0.2467 | No |
| 53 | Plaur | 7057 | -0.091 | -0.2471 | No |
| 54 | Lyn | 7078 | -0.094 | -0.2458 | No |
| 55 | Nfkbia | 7216 | -0.102 | -0.2545 | No |
| 56 | Ripk2 | 7306 | -0.108 | -0.2588 | No |
| 57 | Il15 | 7352 | -0.113 | -0.2592 | No |
| 58 | Ldlr | 7388 | -0.116 | -0.2586 | No |
| 59 | Atp2b1 | 7396 | -0.116 | -0.2555 | No |
| 60 | Nfkb1 | 7406 | -0.117 | -0.2526 | No |
| 61 | Ccrl2 | 7598 | -0.130 | -0.2651 | No |
| 62 | Pik3r5 | 7616 | -0.131 | -0.2624 | No |
| 63 | Gpc3 | 7740 | -0.140 | -0.2687 | No |
| 64 | Icam1 | 7859 | -0.149 | -0.2742 | No |
| 65 | Osm | 8258 | -0.184 | -0.3030 | No |
| 66 | Acvr1b | 8267 | -0.185 | -0.2979 | No |
| 67 | Hrh1 | 8335 | -0.191 | -0.2977 | No |
| 68 | Olr1 | 8449 | -0.198 | -0.3012 | No |
| 69 | Lcp2 | 8504 | -0.205 | -0.2994 | No |
| 70 | Gpr183 | 8553 | -0.209 | -0.2970 | No |
| 71 | Bst2 | 8581 | -0.212 | -0.2927 | No |
| 72 | Cd48 | 8758 | -0.229 | -0.3007 | No |
| 73 | Adgre1 | 8867 | -0.239 | -0.3026 | No |
| 74 | Emp3 | 8874 | -0.240 | -0.2955 | No |
| 75 | Slc11a2 | 9005 | -0.251 | -0.2989 | No |
| 76 | Ebi3 | 9308 | -0.283 | -0.3162 | Yes |
| 77 | Gna15 | 9346 | -0.285 | -0.3104 | Yes |
| 78 | Il18r1 | 9407 | -0.291 | -0.3064 | Yes |
| 79 | Csf3r | 9565 | -0.307 | -0.3103 | Yes |
| 80 | Btg2 | 9641 | -0.315 | -0.3069 | Yes |
| 81 | Hpn | 9704 | -0.322 | -0.3021 | Yes |
| 82 | Kcnj2 | 9738 | -0.327 | -0.2946 | Yes |
| 83 | Ifitm1 | 9752 | -0.329 | -0.2854 | Yes |
| 84 | Klf6 | 9758 | -0.330 | -0.2754 | Yes |
| 85 | Kif1b | 10031 | -0.362 | -0.2876 | Yes |
| 86 | Itga5 | 10047 | -0.363 | -0.2774 | Yes |
| 87 | Raf1 | 10310 | -0.400 | -0.2876 | Yes |
| 88 | Tapbp | 10350 | -0.403 | -0.2782 | Yes |
| 89 | Irf1 | 10434 | -0.415 | -0.2723 | Yes |
| 90 | Abi1 | 10465 | -0.421 | -0.2616 | Yes |
| 91 | Ifngr2 | 10466 | -0.421 | -0.2483 | Yes |
| 92 | Met | 10489 | -0.427 | -0.2367 | Yes |
| 93 | Cmklr1 | 10715 | -0.463 | -0.2417 | Yes |
| 94 | Slc28a2 | 10761 | -0.472 | -0.2307 | Yes |
| 95 | Fzd5 | 10938 | -0.517 | -0.2296 | Yes |
| 96 | Gnai3 | 11081 | -0.555 | -0.2244 | Yes |
| 97 | Cd69 | 11113 | -0.566 | -0.2092 | Yes |
| 98 | P2rx7 | 11131 | -0.571 | -0.1927 | Yes |
| 99 | Csf1 | 11313 | -0.641 | -0.1881 | Yes |
| 100 | Ifnar1 | 11322 | -0.647 | -0.1684 | Yes |
| 101 | Kcna3 | 11449 | -0.735 | -0.1561 | Yes |
| 102 | P2ry2 | 11465 | -0.754 | -0.1336 | Yes |
| 103 | Gabbr1 | 11590 | -1.051 | -0.1111 | Yes |
| 104 | Abca1 | 11595 | -1.077 | -0.0774 | Yes |
| 105 | Ptger4 | 11603 | -1.238 | -0.0389 | Yes |
| 106 | P2rx4 | 11604 | -1.245 | 0.0004 | Yes |