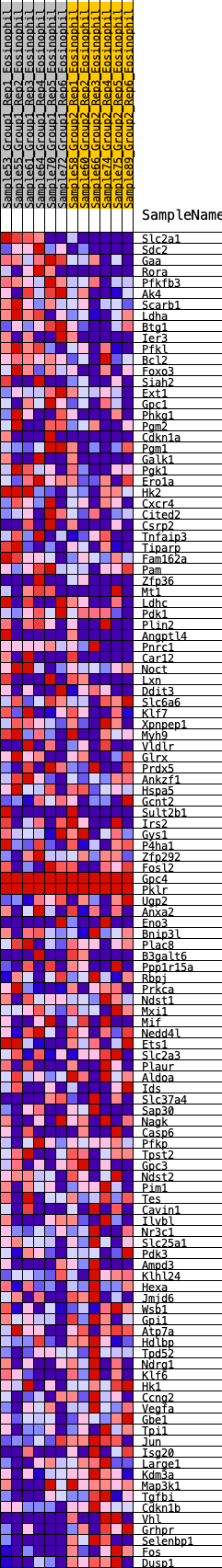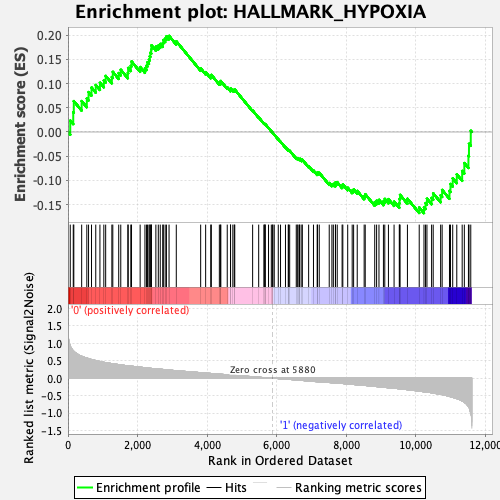
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.19860315 |
| Normalized Enrichment Score (NES) | 0.86347324 |
| Nominal p-value | 0.7077535 |
| FDR q-value | 0.8834016 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc2a1 | 65 | 0.936 | 0.0237 | Yes |
| 2 | Sdc2 | 151 | 0.786 | 0.0409 | Yes |
| 3 | Gaa | 167 | 0.772 | 0.0638 | Yes |
| 4 | Rora | 393 | 0.629 | 0.0639 | Yes |
| 5 | Pfkfb3 | 541 | 0.575 | 0.0691 | Yes |
| 6 | Ak4 | 591 | 0.561 | 0.0824 | Yes |
| 7 | Scarb1 | 676 | 0.534 | 0.0918 | Yes |
| 8 | Ldha | 798 | 0.506 | 0.0972 | Yes |
| 9 | Btg1 | 918 | 0.478 | 0.1018 | Yes |
| 10 | Ier3 | 1030 | 0.455 | 0.1064 | Yes |
| 11 | Pfkl | 1083 | 0.446 | 0.1159 | Yes |
| 12 | Bcl2 | 1261 | 0.418 | 0.1136 | Yes |
| 13 | Foxo3 | 1288 | 0.415 | 0.1243 | Yes |
| 14 | Siah2 | 1458 | 0.389 | 0.1218 | Yes |
| 15 | Ext1 | 1517 | 0.379 | 0.1286 | Yes |
| 16 | Gpc1 | 1721 | 0.354 | 0.1220 | Yes |
| 17 | Phkg1 | 1730 | 0.354 | 0.1324 | Yes |
| 18 | Pgm2 | 1806 | 0.345 | 0.1367 | Yes |
| 19 | Cdkn1a | 1826 | 0.343 | 0.1458 | Yes |
| 20 | Pgm1 | 2074 | 0.316 | 0.1342 | Yes |
| 21 | Galk1 | 2212 | 0.301 | 0.1317 | Yes |
| 22 | Pgk1 | 2258 | 0.297 | 0.1371 | Yes |
| 23 | Ero1a | 2284 | 0.294 | 0.1441 | Yes |
| 24 | Hk2 | 2327 | 0.288 | 0.1495 | Yes |
| 25 | Cxcr4 | 2351 | 0.286 | 0.1565 | Yes |
| 26 | Cited2 | 2368 | 0.283 | 0.1639 | Yes |
| 27 | Csrp2 | 2394 | 0.281 | 0.1705 | Yes |
| 28 | Tnfaip3 | 2398 | 0.280 | 0.1791 | Yes |
| 29 | Tiparp | 2526 | 0.269 | 0.1764 | Yes |
| 30 | Fam162a | 2596 | 0.263 | 0.1787 | Yes |
| 31 | Pam | 2652 | 0.258 | 0.1820 | Yes |
| 32 | Zfp36 | 2724 | 0.251 | 0.1836 | Yes |
| 33 | Mt1 | 2740 | 0.249 | 0.1901 | Yes |
| 34 | Ldhc | 2793 | 0.245 | 0.1933 | Yes |
| 35 | Pdk1 | 2830 | 0.241 | 0.1977 | Yes |
| 36 | Plin2 | 2905 | 0.234 | 0.1986 | Yes |
| 37 | Angptl4 | 3113 | 0.217 | 0.1874 | No |
| 38 | Pnrc1 | 3814 | 0.157 | 0.1314 | No |
| 39 | Car12 | 3957 | 0.145 | 0.1235 | No |
| 40 | Noct | 4104 | 0.136 | 0.1151 | No |
| 41 | Lxn | 4120 | 0.134 | 0.1180 | No |
| 42 | Ddit3 | 4351 | 0.116 | 0.1016 | No |
| 43 | Slc6a6 | 4389 | 0.113 | 0.1019 | No |
| 44 | Klf7 | 4390 | 0.113 | 0.1054 | No |
| 45 | Xpnpep1 | 4578 | 0.100 | 0.0923 | No |
| 46 | Myh9 | 4672 | 0.092 | 0.0871 | No |
| 47 | Vldlr | 4673 | 0.092 | 0.0900 | No |
| 48 | Glrx | 4739 | 0.088 | 0.0871 | No |
| 49 | Prdx5 | 4780 | 0.084 | 0.0862 | No |
| 50 | Ankzf1 | 4794 | 0.082 | 0.0877 | No |
| 51 | Hspa5 | 5308 | 0.043 | 0.0444 | No |
| 52 | Gcnt2 | 5482 | 0.029 | 0.0302 | No |
| 53 | Sult2b1 | 5631 | 0.017 | 0.0179 | No |
| 54 | Irs2 | 5655 | 0.017 | 0.0164 | No |
| 55 | Gys1 | 5665 | 0.016 | 0.0161 | No |
| 56 | P4ha1 | 5668 | 0.016 | 0.0165 | No |
| 57 | Zfp292 | 5765 | 0.009 | 0.0084 | No |
| 58 | Fosl2 | 5845 | 0.003 | 0.0016 | No |
| 59 | Gpc4 | 5883 | 0.000 | -0.0016 | No |
| 60 | Pklr | 5888 | 0.000 | -0.0020 | No |
| 61 | Ugp2 | 5929 | -0.002 | -0.0054 | No |
| 62 | Anxa2 | 6045 | -0.011 | -0.0151 | No |
| 63 | Eno3 | 6110 | -0.017 | -0.0201 | No |
| 64 | Bnip3l | 6252 | -0.026 | -0.0315 | No |
| 65 | Plac8 | 6334 | -0.031 | -0.0376 | No |
| 66 | B3galt6 | 6336 | -0.032 | -0.0367 | No |
| 67 | Ppp1r15a | 6373 | -0.035 | -0.0388 | No |
| 68 | Rbpj | 6562 | -0.050 | -0.0536 | No |
| 69 | Prkca | 6591 | -0.052 | -0.0544 | No |
| 70 | Ndst1 | 6615 | -0.054 | -0.0547 | No |
| 71 | Mxi1 | 6656 | -0.058 | -0.0563 | No |
| 72 | Mif | 6666 | -0.059 | -0.0553 | No |
| 73 | Nedd4l | 6725 | -0.064 | -0.0583 | No |
| 74 | Ets1 | 6734 | -0.065 | -0.0570 | No |
| 75 | Slc2a3 | 6918 | -0.080 | -0.0704 | No |
| 76 | Plaur | 7057 | -0.091 | -0.0796 | No |
| 77 | Aldoa | 7164 | -0.099 | -0.0857 | No |
| 78 | Ids | 7172 | -0.099 | -0.0832 | No |
| 79 | Slc37a4 | 7223 | -0.103 | -0.0843 | No |
| 80 | Sap30 | 7510 | -0.123 | -0.1054 | No |
| 81 | Nagk | 7587 | -0.130 | -0.1079 | No |
| 82 | Casp6 | 7628 | -0.131 | -0.1073 | No |
| 83 | Pfkp | 7688 | -0.136 | -0.1082 | No |
| 84 | Tpst2 | 7690 | -0.136 | -0.1040 | No |
| 85 | Gpc3 | 7740 | -0.140 | -0.1039 | No |
| 86 | Ndst2 | 7880 | -0.151 | -0.1112 | No |
| 87 | Pim1 | 7901 | -0.152 | -0.1082 | No |
| 88 | Tes | 8041 | -0.164 | -0.1152 | No |
| 89 | Cavin1 | 8170 | -0.175 | -0.1208 | No |
| 90 | Ilvbl | 8211 | -0.179 | -0.1187 | No |
| 91 | Nr3c1 | 8315 | -0.189 | -0.1217 | No |
| 92 | Slc25a1 | 8514 | -0.206 | -0.1325 | No |
| 93 | Pdk3 | 8545 | -0.208 | -0.1286 | No |
| 94 | Ampd3 | 8818 | -0.233 | -0.1449 | No |
| 95 | Klhl24 | 8872 | -0.240 | -0.1420 | No |
| 96 | Hexa | 8940 | -0.247 | -0.1401 | No |
| 97 | Jmjd6 | 9068 | -0.258 | -0.1431 | No |
| 98 | Wsb1 | 9107 | -0.262 | -0.1382 | No |
| 99 | Gpi1 | 9212 | -0.273 | -0.1387 | No |
| 100 | Atp7a | 9374 | -0.288 | -0.1437 | No |
| 101 | Hdlbp | 9524 | -0.302 | -0.1472 | No |
| 102 | Tpd52 | 9532 | -0.303 | -0.1383 | No |
| 103 | Ndrg1 | 9546 | -0.305 | -0.1299 | No |
| 104 | Klf6 | 9758 | -0.330 | -0.1379 | No |
| 105 | Hk1 | 10097 | -0.368 | -0.1558 | No |
| 106 | Ccng2 | 10234 | -0.389 | -0.1554 | No |
| 107 | Vegfa | 10279 | -0.395 | -0.1469 | No |
| 108 | Gbe1 | 10319 | -0.401 | -0.1377 | No |
| 109 | Tpi1 | 10453 | -0.419 | -0.1362 | No |
| 110 | Jun | 10499 | -0.428 | -0.1267 | No |
| 111 | Isg20 | 10713 | -0.463 | -0.1307 | No |
| 112 | Large1 | 10756 | -0.470 | -0.1197 | No |
| 113 | Kdm3a | 10967 | -0.523 | -0.1216 | No |
| 114 | Map3k1 | 10996 | -0.530 | -0.1074 | No |
| 115 | Tgfbi | 11060 | -0.549 | -0.0957 | No |
| 116 | Cdkn1b | 11177 | -0.584 | -0.0875 | No |
| 117 | Vhl | 11332 | -0.653 | -0.0804 | No |
| 118 | Grhpr | 11394 | -0.688 | -0.0642 | No |
| 119 | Selenbp1 | 11512 | -0.811 | -0.0490 | No |
| 120 | Fos | 11528 | -0.831 | -0.0242 | No |
| 121 | Dusp1 | 11579 | -0.995 | 0.0026 | No |