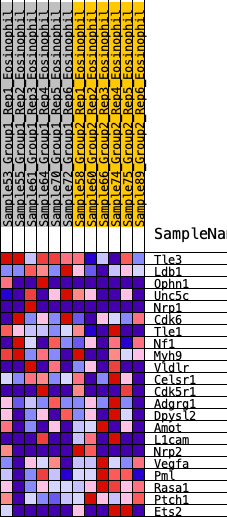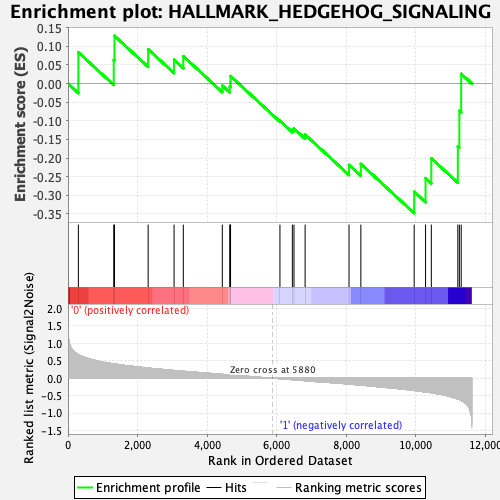
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | -0.34793082 |
| Normalized Enrichment Score (NES) | -1.1213591 |
| Nominal p-value | 0.32149902 |
| FDR q-value | 0.47664988 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tle3 | 298 | 0.673 | 0.0840 | No |
| 2 | Ldb1 | 1315 | 0.411 | 0.0633 | No |
| 3 | Ophn1 | 1337 | 0.408 | 0.1281 | No |
| 4 | Unc5c | 2304 | 0.291 | 0.0922 | No |
| 5 | Nrp1 | 3049 | 0.221 | 0.0640 | No |
| 6 | Cdk6 | 3315 | 0.199 | 0.0736 | No |
| 7 | Tle1 | 4436 | 0.110 | -0.0052 | No |
| 8 | Nf1 | 4649 | 0.094 | -0.0081 | No |
| 9 | Myh9 | 4672 | 0.092 | 0.0050 | No |
| 10 | Vldlr | 4673 | 0.092 | 0.0201 | No |
| 11 | Celsr1 | 6093 | -0.015 | -0.0999 | No |
| 12 | Cdk5r1 | 6450 | -0.040 | -0.1241 | No |
| 13 | Adgrg1 | 6497 | -0.044 | -0.1208 | No |
| 14 | Dpysl2 | 6817 | -0.071 | -0.1369 | No |
| 15 | Amot | 8079 | -0.167 | -0.2185 | No |
| 16 | L1cam | 8419 | -0.196 | -0.2157 | No |
| 17 | Nrp2 | 9952 | -0.353 | -0.2904 | Yes |
| 18 | Vegfa | 10279 | -0.395 | -0.2541 | Yes |
| 19 | Pml | 10445 | -0.416 | -0.2005 | Yes |
| 20 | Rasa1 | 11207 | -0.599 | -0.1686 | Yes |
| 21 | Ptch1 | 11251 | -0.611 | -0.0726 | Yes |
| 22 | Ets2 | 11304 | -0.634 | 0.0263 | Yes |